For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDSFNPTTKTQAALTSALQAASAAGNPEIRPAHLLMALLTQADGIAAPLLEAVGVDPATIRTEAQRLVDR LPQASGASTQPQLSRESLAAITTAQQLATEMDDEYVSTEHVLVGLATGDSEVAKLLTGHGASPQALREAF VKVRGSARVTSPEPEATYQALEKYSTDLTARAREGKLDPVIGRDNEIRRVVQVLSRRTKNNPVLIGEPGV GKTAIVEGLAQRIVAGDVPESLRDKTIVALDLGSMVAGSKYRGEFEERLKAVLDDIKNSAGQIITFIDEL HTIVGAGATGEGAMDAGNMIKPMLARGELRLVGATTLDEYRKHIEKDAALERRFQQVYVGEPSAEDTIGI LRGLKDRYEVHHGVRITDSALVAAATLSDRYITARFLPDKAIDLVDEAASRLRMEIDSRPVEIDEVERLV RRLEIEEMALSKEEDDASKDRLEKLRAELADKKEELAELTTRWQNEKNSIEIVRELKEQLDALRGESERA ERDGDLAKAAELRYGRIPEMEKKLDAAVPHAQAREQVMLKEEVGPDDIAEVVSAWTGIPAGRMLEGETAK LLRMEDELGHRVIGQKKAVQAVSDAVRRSRAGVADPNRPTGSFMFLGPTGVGKTELAKALAEFLFDDERA MVRIDMSEYGEKHSVARLVGAPPGYIGYDQGGQLTEAVRRRPYTVILFDEIEKAHPDVFDVLLQVLDEGR LTDGQGRTVDFRNTILILTSNLGSGGTPDQVMAAVRAAFKPEFINRLDDVLIFDALNPDELVQIVDIQLQ QLDKRLAQRRLQLEVSLPAKEWLAQRGFDPVYGARPLRRLVQQAIGDQLAKMLLAGQVHDGDTVPVNVSA DGESLVLG
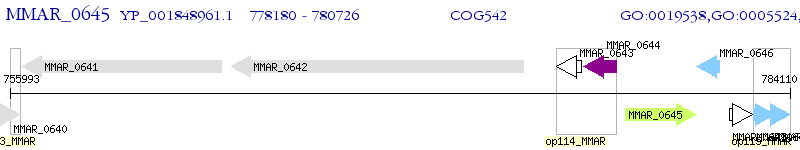
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0645 | clpB | - | 100% (848) | endopeptidase ATP binding protein (chain B) ClpB |
| M. marinum M | MMAR_5100 | clpC1 | 0.0 | 47.13% (872) | ATP-dependent protease ATP-binding subunit ClpC1 |
| M. marinum M | MMAR_1785 | smc | 9e-05 | 24.76% (206) | chromosome partition protein Smc |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0391c | clpB | 0.0 | 93.04% (848) | endopeptidase ATP-binding protein ClpB |
| M. gilvum PYR-GCK | Mflv_0246 | - | 0.0 | 90.45% (848) | ATPase |
| M. tuberculosis H37Rv | Rv0384c | clpB | 0.0 | 93.04% (848) | endopeptidase ATP binding protein |
| M. leprae Br4923 | MLBr_02490 | clpB | 0.0 | 88.92% (848) | heat shock protein |
| M. abscessus ATCC 19977 | MAB_4265c | - | 0.0 | 86.44% (848) | chaperone ClpB |
| M. avium 104 | MAV_4793 | - | 0.0 | 92.57% (848) | chaperone ClpB |
| M. smegmatis MC2 155 | MSMEG_0732 | - | 0.0 | 92.81% (848) | chaperone ClpB |
| M. thermoresistible (build 8) | TH_2647 | - | 0.0 | 88.21% (848) | PUTATIVE chaperone ClpB |
| M. ulcerans Agy99 | MUL_0127 | clpB | 0.0 | 99.76% (848) | endopeptidase ATP binding protein (chain B) ClpB |
| M. vanbaalenii PYR-1 | Mvan_0659 | - | 0.0 | 89.98% (848) | ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0246|M.gilvum_PYR-GCK ------MDSFNPTTKTQAALTSALQAATAAGNPQITPAHLLMALLTQNDG
Mvan_0659|M.vanbaalenii_PYR-1 ------MDSFNPTTKTQAALTSALQAATAAGNPQIAPAHLLMALLTQNDG
MSMEG_0732|M.smegmatis_MC2_155 ------MDSFNPTTKTQAALTSALQAASSAGNPEIRPAHLLMALLTQNDG
TH_2647|M.thermoresistible__bu LTIGGVVDSFNPTTKTQAALTAALQAATAAGHPQIQPAHLLMALLTQHDG
MMAR_0645|M.marinum_M ------MDSFNPTTKTQAALTSALQAASAAGNPEIRPAHLLMALLTQADG
MUL_0127|M.ulcerans_Agy99 ------MDSFNPTTKTQAALTSALQAASAAGNPEIRPAHLLMALLTQADG
Mb0391c|M.bovis_AF2122/97 ------MDSFNPTTKTQAALTAALQAASTAGNPEIRPAHLLMALLTQNDG
Rv0384c|M.tuberculosis_H37Rv ------MDSFNPTTKTQAALTAALQAASTAGNPEIRPAHLLMALLTQNDG
MLBr_02490|M.leprae_Br4923 ------MDSFNPTTKMQVALTSALQAASSAGNPEIRPVHLLLAMLMQNDG
MAV_4793|M.avium_104 ------MDSFNPTTKTQAALTAALQAASAAGNPEIRPAHLLMALLTQADG
MAB_4265c|M.abscessus_ATCC_199 ------MDSFNPTTKTQAALTAALQAATTAGNPEIRPAHLLVALLGQTDG
:******** *.***:*****::**:*:* *.***:*:* * **
Mflv_0246|M.gilvum_PYR-GCK IAAPLLEAVGVEPATIRAETQRLLDRLPSATGSASQPQLSPPAIAAITAA
Mvan_0659|M.vanbaalenii_PYR-1 IAAPLLEAVGVEPATIRAETQRLLDRLPSATGSSSQPQLAPQAIAAITAA
MSMEG_0732|M.smegmatis_MC2_155 IAAPLLEAVGVEPATIRTETQRLLDRLPSASGASSQPQLSRESLAAITTA
TH_2647|M.thermoresistible__bu IAAPLLEAVGVEPATIRAEVQRVLDQQPKISGGASQPQLSRDSLNAITTA
MMAR_0645|M.marinum_M IAAPLLEAVGVDPATIRTEAQRLVDRLPQASGASTQPQLSRESLAAITTA
MUL_0127|M.ulcerans_Agy99 IAAPLLEAVGVDPATIRTEAQRLVDRLPQASGASTQPQLSRESLAAITTA
Mb0391c|M.bovis_AF2122/97 IAAPLLEAVGVEPATVRAETQRLLDRLPQATGASTQPQLSRESLAAITTA
Rv0384c|M.tuberculosis_H37Rv IAAPLLEAVGVEPATVRAETQRLLDRLPQATGASTQPQLSRESLAAITTA
MLBr_02490|M.leprae_Br4923 IAAPLLETVGVEPDIVRNEAQRLLERLPQASGCSSQPQLSRESLAAITTA
MAV_4793|M.avium_104 IAAPLLEAVGVEPATIRAEAERMLARLPQASGASSQPQLSRESLAAITTA
MAB_4265c|M.abscessus_ATCC_199 IAAPLLQAVGVDPVSVRNEAQAIADRLPQVSNASANPQLSRDSIAAVTAA
******::***:* :* *.: : : *. :. :::***: :: *:*:*
Mflv_0246|M.gilvum_PYR-GCK THLATEMDDEYVSTEHLLVGLATGDSDVAKVLTGHGASPQALREAFVKVR
Mvan_0659|M.vanbaalenii_PYR-1 THLASEMDDEYVSTEHLLVGLATGDADTAKLLTGHGASPQALREAFVKVR
MSMEG_0732|M.smegmatis_MC2_155 QNLATEMDDEYVSTEHLMVGLATGDSEVAKLLTGHGASPQALREAFVKVR
TH_2647|M.thermoresistible__bu QQLATEMADEYVSTEHLLVGLASGDSDVAKILNGHGATPQALREAFAKVR
MMAR_0645|M.marinum_M QQLATEMDDEYVSTEHVLVGLATGDSEVAKLLTGHGASPQALREAFVKVR
MUL_0127|M.ulcerans_Agy99 QQLATEMDDEYVSTEHVLVGLATGDSEVAKLLTGHGASPQALREAFVKVR
Mb0391c|M.bovis_AF2122/97 QQLATELDDEYVSTEHVMVGLATGDSDVAKLLTGHGASPQALREAFVKVR
Rv0384c|M.tuberculosis_H37Rv QQLATELDDEYVSTEHVMVGLATGDSDVAKLLTGHGASPQALREAFVKVR
MLBr_02490|M.leprae_Br4923 QQLATEMDDEYVSTEHLMLGLAMGDSDVAKLLTGHGASPQVLREAFVKIR
MAV_4793|M.avium_104 QHLATELDDEYVSTEHLMVGLATGDSDVAKLLTGHGASPQALREAFVKVR
MAB_4265c|M.abscessus_ATCC_199 QHLATELNDDYVSTEHLLVGLATGDSDIAKLLVNHGATPQALRDAFVQVR
:**:*: *:******:::*** **:: **:* .***:**.**:**.::*
Mflv_0246|M.gilvum_PYR-GCK GSARVTNPDPEGTYQALEKYSTDLTARAREGKLDPVIGRDHEIRRVVQVL
Mvan_0659|M.vanbaalenii_PYR-1 GSARVTSPDPEGTYQALEKYSTDLTARAREGKLDPVIGRDNEIRRVVQVL
MSMEG_0732|M.smegmatis_MC2_155 GSARVTSPDPEGTYQALEKYSTDLTARAREGKLDPVIGRDNEIRRVVQVL
TH_2647|M.thermoresistible__bu GSARVTTPDPEGTYQALEKYSTDLTARAREGKLDPVIGRDNEIRRVIQVL
MMAR_0645|M.marinum_M GSARVTSPEPEATYQALEKYSTDLTARAREGKLDPVIGRDNEIRRVVQVL
MUL_0127|M.ulcerans_Agy99 GSARVTSPEPEATYQALEKYSTDLTARAREGKLDPVIGRDNEIRRVVQVL
Mb0391c|M.bovis_AF2122/97 GSARVTSPEPEATYQALQKYSTDLTARAREGKLDPVIGRDNEIRRVVQVL
Rv0384c|M.tuberculosis_H37Rv GSARVTSPEPEATYQALQKYSTDLTARAREGKLDPVIGRDNEIRRVVQVL
MLBr_02490|M.leprae_Br4923 GSARVTSSDPEFTYQALEKYSTDLTAQSGEGKLDPVIGRDNEIRRVVQVL
MAV_4793|M.avium_104 GSARVTSPDPEATYQALEKYSTDLTARAREGKLDPVIGRDNEIRRVVQVL
MAB_4265c|M.abscessus_ATCC_199 GSGRVTSPEPEATFQALEKYSTDLTARAREGKLDPVIGRDTEIRRVVQVL
**.***..:** *:***:********:: *********** *****:***
Mflv_0246|M.gilvum_PYR-GCK SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTVVSLDMGS
Mvan_0659|M.vanbaalenii_PYR-1 SRRTKNNPVLIGEPGVGKTAIVEGLAQRIIAGDVPESLRDKTVVSLDLGS
MSMEG_0732|M.smegmatis_MC2_155 SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTVIALDLGS
TH_2647|M.thermoresistible__bu SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTVIALDLGS
MMAR_0645|M.marinum_M SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTIVALDLGS
MUL_0127|M.ulcerans_Agy99 SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTIVALDLGS
Mb0391c|M.bovis_AF2122/97 SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTIVALDLGS
Rv0384c|M.tuberculosis_H37Rv SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTIVALDLGS
MLBr_02490|M.leprae_Br4923 SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTLVALDLGS
MAV_4793|M.avium_104 SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRDKTVIALDLGS
MAB_4265c|M.abscessus_ATCC_199 SRRTKNNPVLIGEPGVGKTAIVEGLAQRIVAGDVPESLRGKTVISLDLGS
*****************************:*********.**:::**:**
Mflv_0246|M.gilvum_PYR-GCK MVAGAKYRGEFEERLKAVLDDIKNSAGQIITFIDELHTIVGAGATGESAM
Mvan_0659|M.vanbaalenii_PYR-1 MVAGAKYRGEFEERLKAVLDDIKNSAGQIITFIDELHTIVGAGATGESAM
MSMEG_0732|M.smegmatis_MC2_155 MVAGAKYRGEFEERLKAVLDDIKNSAGQVITFIDELHTIVGAGATGESAM
TH_2647|M.thermoresistible__bu MVAGSKYRGEFEERLKAVLDEIKNSAGQIITFIDELHTIVGAGATGDGSM
MMAR_0645|M.marinum_M MVAGSKYRGEFEERLKAVLDDIKNSAGQIITFIDELHTIVGAGATGEGAM
MUL_0127|M.ulcerans_Agy99 MVAGSKYRGEFEERLKAVLDDIKNSAGQIITFIDELHTIVGAGATGEGAM
Mb0391c|M.bovis_AF2122/97 MVAGSKYRGEFEERLKAVLDDIKNSAGQIITFIDELHTIVGAGATGEGAM
Rv0384c|M.tuberculosis_H37Rv MVAGSKYRGEFEERLKAVLDDIKNSAGQIITFIDELHTIVGAGATGEGAM
MLBr_02490|M.leprae_Br4923 MVAGAKYRGEFEQRLKVVLDDIKFSAGQIITFIDELHTIVGAGATGESAM
MAV_4793|M.avium_104 MVAGAKYRGEFEERLKAVLDDIKNSAGQIITFIDELHTIVGAGATGEGAM
MAB_4265c|M.abscessus_ATCC_199 MVAGAKYRGEFEERLKAVLDEIKNSAGQLITFIDELHTIVGAGATGESAM
****:*******:***.***:** ****:*****************:.:*
Mflv_0246|M.gilvum_PYR-GCK DAGNMIKPMLARGELRLVGATTLDEYRKYIEKDAALERRFQQVLVGEPSV
Mvan_0659|M.vanbaalenii_PYR-1 DAGNMIKPMLARGELRLVGATTLDEYRKYIEKDAALERRFQQVMVGEPSV
MSMEG_0732|M.smegmatis_MC2_155 DAGNMIKPMLARGELRLVGATTLDEYRKYIEKDAALERRFQQVFVGEPSV
TH_2647|M.thermoresistible__bu DAGNMIKPMLARGELRLVGATTLDEYRKYIEKDAALERRFQQVYVGEPSV
MMAR_0645|M.marinum_M DAGNMIKPMLARGELRLVGATTLDEYRKHIEKDAALERRFQQVYVGEPSA
MUL_0127|M.ulcerans_Agy99 DAGNMIKPMLARGELRLVGATTLDEYRKHIEKDAALERRFQQVYVGEPSA
Mb0391c|M.bovis_AF2122/97 DAGNMIKPMLARGELRLVGATTLDEYRKHIEKDAALERRFQQVYVGEPSV
Rv0384c|M.tuberculosis_H37Rv DAGNMIKPMLARGELRLVGATTLDEYRKHIEKDAALERRFQQVYVGEPSV
MLBr_02490|M.leprae_Br4923 DAGNMIKPMLARGELRLVGATTLDEYRRYIEKDAALERRFQQIFVGEPSV
MAV_4793|M.avium_104 DAGNMIKPMLARGELRLVGATTLDEYRKYIEKDAALERRFQQVFVGEPSV
MAB_4265c|M.abscessus_ATCC_199 DAGNMIKPMLARGELRLVGATTLDEYRKYIEKDAALERRFQQVLVGEPSV
***************************::*************: *****.
Mflv_0246|M.gilvum_PYR-GCK EDTVGILRGLKDRYEVHHGVRITDSSLVAAATLSDRYITSRFLPDKAIDL
Mvan_0659|M.vanbaalenii_PYR-1 EDTVGILRGLKERYEVHHGVRITDSALVSAATLSDRYITARFLPDKAIDL
MSMEG_0732|M.smegmatis_MC2_155 EDTVGILRGLKDRYEVHHGVRITDSALVAAATLSDRYITARFLPDKAIDL
TH_2647|M.thermoresistible__bu EDTIGILRGLKERYEVHHGVRITDSALVAAATLSNRYITARFLPDKAIDL
MMAR_0645|M.marinum_M EDTIGILRGLKDRYEVHHGVRITDSALVAAATLSDRYITARFLPDKAIDL
MUL_0127|M.ulcerans_Agy99 EDTIGILRGLKDRYEVHHGVRITDSALVAAATLSDRYITARFLPDKAIDL
Mb0391c|M.bovis_AF2122/97 EDTIGILRGLKDRYEVHHGVRITDSALVAAATLSDRYITARFLPDKAIDL
Rv0384c|M.tuberculosis_H37Rv EDTIGILRGLKDRYEVHHGVRITDSALVAAATLSDRYITARFLPDKAIDL
MLBr_02490|M.leprae_Br4923 EDTVGILRGLKDRYEVHHGVRITDSALVAAAALSDRYITARFLPDKAIDL
MAV_4793|M.avium_104 EDTVGILRGLKDRYEVHHGVRITDSALVAAATLSDRYITARFLPDKAIDL
MAB_4265c|M.abscessus_ATCC_199 EDTIGILRGIKERYEIHHGVRITDSALVAAATLSDRYITSRFLPDKAIDL
***:*****:*:***:*********:**:**:**:****:**********
Mflv_0246|M.gilvum_PYR-GCK VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALAKEEDAASKDRLEK
Mvan_0659|M.vanbaalenii_PYR-1 VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALEKESDAASKDRLEK
MSMEG_0732|M.smegmatis_MC2_155 VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALEKEEDEASKDRLEK
TH_2647|M.thermoresistible__bu VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALSKEEDEASKERLEK
MMAR_0645|M.marinum_M VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALSKEEDDASKDRLEK
MUL_0127|M.ulcerans_Agy99 VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALSKEEDDASKDRLEK
Mb0391c|M.bovis_AF2122/97 VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALSKEEDEASAERLAK
Rv0384c|M.tuberculosis_H37Rv VDEAASRLRMEIDSRPVEIDEVERLVRRLEIEEMALSKEEDEASAERLAK
MLBr_02490|M.leprae_Br4923 VDEAASRLRMEIDSRPVEIDEVERLVRRFEIEEMALVNEEDEASKERLET
MAV_4793|M.avium_104 VDEAASRLKMEIDSRPVEIDEVERLVRRLEIEEMALAKEEDEASKERLEK
MAB_4265c|M.abscessus_ATCC_199 VDEAASRLRMEIDSRPVEIDEVERVVRRLEIEEMALSKEEDEASKQRLEK
********:***************:***:******* :*.* ** :** .
Mflv_0246|M.gilvum_PYR-GCK LRAELADKKEQLAELTTRWQNEKSAIDVVRELKEQLESLRGEADRAERDG
Mvan_0659|M.vanbaalenii_PYR-1 LRSELADQKEKLSELTTRWQNEKGAIDVVRELKEQLEELRGAADRAERDG
MSMEG_0732|M.smegmatis_MC2_155 LRAELADHKEKLSELTTRWQNEKNAINVVRELKEQLDTLRGEADRAERDG
TH_2647|M.thermoresistible__bu LRAELADHKEKLAELTARWQNEKKAIETVRELKEQLEALKIEAERAERDG
MMAR_0645|M.marinum_M LRAELADKKEELAELTTRWQNEKNSIEIVRELKEQLDALRGESERAERDG
MUL_0127|M.ulcerans_Agy99 LRAELADKKEELAELTTRWQNEKNSIEIVRELKEQLDALRGESERAERDG
Mb0391c|M.bovis_AF2122/97 LRSELADQKEKLAELTTRWQNEKNAIEIVRDLKEQLEALRGESERAERDG
Rv0384c|M.tuberculosis_H37Rv LRSELADQKEKLAELTTRWQNEKNAIEIVRDLKEQLEALRGESERAERDG
MLBr_02490|M.leprae_Br4923 LRAELADQKERLAELTARWQNEKNAIDVVRELKEQLETLRGESERAERDG
MAV_4793|M.avium_104 LRSELADQKEKLAELTTRWQNEKNAIDVVRELKEQLETLRGESDRAERDG
MAB_4265c|M.abscessus_ATCC_199 LRVELADKKERLAELTARWQNEKSSIDAVRDLKEQLETLKGESDRAERDG
** ****:**.*:***:****** :*: **:*****: *: ::******
Mflv_0246|M.gilvum_PYR-GCK DLAKAAELRYGRIPEVEKKLDAALPHAEARENVMLKEEVGPDDIAEVVEA
Mvan_0659|M.vanbaalenii_PYR-1 DLAKAAELRYGRIPEVEKKLDAAVPQAEARDNVMLKEEVGPDDIADVVSA
MSMEG_0732|M.smegmatis_MC2_155 DLAKAAELRYGRIPEVEKKLDAALPAAEARENLMLKEEVGPDDIAEVVEA
TH_2647|M.thermoresistible__bu NYERVAELRYGRIPEVEKKLEAATPHAEARENVMLKEEVGPDDIAEVVSA
MMAR_0645|M.marinum_M DLAKAAELRYGRIPEMEKKLDAAVPHAQAREQVMLKEEVGPDDIAEVVSA
MUL_0127|M.ulcerans_Agy99 DLAKAAELRYGRIPEMEKKLDAAVPHAQAREQVMLKEEVGPDDIAEVVSA
Mb0391c|M.bovis_AF2122/97 DLAKAAELRYGRIPEVEKKLDAALPQAQAREQVMLKEEVGPDDIADVVSA
Rv0384c|M.tuberculosis_H37Rv DLAKAAELRYGRIPEVEKKLDAALPQAQAREQVMLKEEVGPDDIADVVSA
MLBr_02490|M.leprae_Br4923 DLAKAAELRYGRIPEVEKKLEAAVPTAQAREDVMLKEEVGPDDIANVVSA
MAV_4793|M.avium_104 DLAKAAELRYGRIPEVEKKLEAALPQAEARENVMLKEEVGPDDIAEVVSA
MAB_4265c|M.abscessus_ATCC_199 DLGKAAELRYGRIPELEKQLEQALPGLDHDGNVMLKEEVSPDDVADVVSA
: :.**********:**:*: * * : ::******.***:*:**.*
Mflv_0246|M.gilvum_PYR-GCK WTGIPAGRMLEGETAKLLRMEEELGKRVVGQRKAVTAVSDAVRRSRAGVA
Mvan_0659|M.vanbaalenii_PYR-1 WTGIPAGRMLEGETAKLLRMEDELGKRVVGQKNAVQAVSDAVRRSRAGVA
MSMEG_0732|M.smegmatis_MC2_155 WTGIPSGRMLEGETAKLLRMEEELGKRVVGQRKAVQAVSDAVRRSRAGVA
TH_2647|M.thermoresistible__bu WTGIPAGRMLEGEQAKLLRMEDELAKRVVGQKRAVQAVSDAVRRSRAGVA
MMAR_0645|M.marinum_M WTGIPAGRMLEGETAKLLRMEDELGHRVIGQKKAVQAVSDAVRRSRAGVA
MUL_0127|M.ulcerans_Agy99 WTGIPAGRMLEGETAKLLRMEDELGHRVIGQKKAVQAVSDAVRRSRAGVA
Mb0391c|M.bovis_AF2122/97 WTGIPAGRLLEGETAKLLRMEDELGKRVIGQKAAVTAVSDAVRRSRAGVS
Rv0384c|M.tuberculosis_H37Rv WTGIPAGRLLEGETAKLLRMEDELGKRVIGQKAAVTAVSDAVRRSRAGVS
MLBr_02490|M.leprae_Br4923 WTGIPAGRLLEGETAKLLRMEDELGKRVVGQKKAVHAVSDAVRRSRAGVA
MAV_4793|M.avium_104 WTGIPAGRMLEGETAKLLRMEDELGKRVVGQKRAVQAVSDAVRRARAGVA
MAB_4265c|M.abscessus_ATCC_199 WTGIPTGRLMEGETAKLLRMEDELGKRVVGQKKAVEAVSDAVRRARAGVA
*****:**::*** *******:**.:**:**: ** ********:****:
Mflv_0246|M.gilvum_PYR-GCK DPNRPTGSFMFLGPTGVGKTELAKALAEFLFDDERAMVRIDMSEYGEKHS
Mvan_0659|M.vanbaalenii_PYR-1 DPNRPTGSFMFLGPTGVGKTELAKALAEFLFDDERAMVRIDMSEYGEKHS
MSMEG_0732|M.smegmatis_MC2_155 DPNRPTGSFMFLGPTGVGKTELAKALAEFLFDDERAMVRIDMSEYGEKHS
TH_2647|M.thermoresistible__bu DPNRPTGSFMFLGPTGVGKTELAKALAEFLFDDERAMVRIDMSEYSEKHS
MMAR_0645|M.marinum_M DPNRPTGSFMFLGPTGVGKTELAKALAEFLFDDERAMVRIDMSEYGEKHS
MUL_0127|M.ulcerans_Agy99 DPNRPTGSFMFLGPTGVGKTELAKALAEFLFDDERAMVRIDMSEYGEKHS
Mb0391c|M.bovis_AF2122/97 DPNRPTGAFMFLGPTGVGKTELAKALADFLFDDERAMVRIDMSEYGEKHT
Rv0384c|M.tuberculosis_H37Rv DPNRPTGAFMFLGPTGVGKTELAKALADFLFDDERAMVRIDMSEYGEKHT
MLBr_02490|M.leprae_Br4923 DPNRPTGSFLFLGPTGVGKTELAKALADFLFDDQRAMVRVDMSEYGEKHS
MAV_4793|M.avium_104 DPNRPTGSFMFLGPTGVGKTELAKALADFLFDDERAMVRIDMSEYGEKHS
MAB_4265c|M.abscessus_ATCC_199 DPNRPTGSFLFLGPTGVGKTELAKALADFLFDDEHAMVRIDMSEYGEKHS
*******:*:*****************:*****::****:*****.***:
Mflv_0246|M.gilvum_PYR-GCK VARLVGAPPGYVGYDQGGQLTEAVRRRPYTVILFDEIEKAHPDVFDVLLQ
Mvan_0659|M.vanbaalenii_PYR-1 VARLVGAPPGYVGYDQGGQLTEAVRRRPYTVILFDEIEKAHPDVFDVLLQ
MSMEG_0732|M.smegmatis_MC2_155 VARLVGAPPGYIGYDQGGQLTEAVRRRPYTVILFDEIEKAHPDVFDVLLQ
TH_2647|M.thermoresistible__bu VARLVGAPPGYVGYDQGGQLTEAVRRRPYTVILFDEIEKAHQDVFDILLQ
MMAR_0645|M.marinum_M VARLVGAPPGYIGYDQGGQLTEAVRRRPYTVILFDEIEKAHPDVFDVLLQ
MUL_0127|M.ulcerans_Agy99 VARLVGAPPGYIGYDHGGQLTEAVRRRPYTVILFDEIEKAHPDVFDVLLQ
Mb0391c|M.bovis_AF2122/97 VARLIGAPPGYVGYEAGGQLTEAVRRRPYTVVLFDEIEKAHPDVFDVLLQ
Rv0384c|M.tuberculosis_H37Rv VARLIGAPPGYVGYEAGGQLTEAVRRRPYTVVLFDEIEKAHPDVFDVLLQ
MLBr_02490|M.leprae_Br4923 VARLVGAPPGYIGHDQGGQLTEAVRRRPYTVVLFDEVEKAHSDVFDVLLQ
MAV_4793|M.avium_104 VARLVGAPPGYIGYDQGGQLTEAVRRRPYTVILFDEIEKAHPDVFDVLLQ
MAB_4265c|M.abscessus_ATCC_199 VARLVGAPPGYVGYDAGGQLTEAVRRRPYTVVLFDEVEKAHPDVFDVLLQ
****:******:*:: ***************:****:**** ****:***
Mflv_0246|M.gilvum_PYR-GCK VLDEGRLTDGQGRTVDFRNTILILTSNLGAGGSEEQVMAAVRAAFKPEFI
Mvan_0659|M.vanbaalenii_PYR-1 VLDEGRLTDGQGRTVDFRNTILILTSNLGAGGNEEQVMAAVRAAFKPEFI
MSMEG_0732|M.smegmatis_MC2_155 VLDEGRLTDGQGRTVDFRNTILILTSNLGAGGNEDQVMAAVRSAFKPEFI
TH_2647|M.thermoresistible__bu VLDDGRLTDGQGRTVDFRNTILILTSNLGAGGSEEQVMAAVRAHFKPEFI
MMAR_0645|M.marinum_M VLDEGRLTDGQGRTVDFRNTILILTSNLGSGGTPDQVMAAVRAAFKPEFI
MUL_0127|M.ulcerans_Agy99 VLDEGRLTDGQGRTVDFRNTILILTSNLGSGGTPDQVMAAVRAAFKPEFI
Mb0391c|M.bovis_AF2122/97 VLDEGRLTDGHGRTVDFRNTILILTSNLGSGGSAEQVLAAVRATFKPEFI
Rv0384c|M.tuberculosis_H37Rv VLDEGRLTDGHGRTVDFRNTILILTSNLGSGGSAEQVLAAVRATFKPEFI
MLBr_02490|M.leprae_Br4923 VLDEGRLTDGQGRTVDFRNTIPILTSNLGSGGSEEQVMAAVRSVFKPEFI
MAV_4793|M.avium_104 VLDEGRLTDGQGRTVDFRNTILILTSNLGSGGSEEQVMAAVRSAFKPEFI
MAB_4265c|M.abscessus_ATCC_199 VLDEGRLTDGQGRTVDFRNTILILTSNLGAGGSEEQVMAAVRAKFKPEFI
***:******:********** *******:**. :**:****: ******
Mflv_0246|M.gilvum_PYR-GCK NRLDDVIMFDALDPEQLVSIVDIQLEQLAKRLAQRRLTLEVSLPAKKWLA
Mvan_0659|M.vanbaalenii_PYR-1 NRLDDVIMFDALNPEQLVSIVDIQLAQLQKRLAQRRLTLEVSLPAKKWLA
MSMEG_0732|M.smegmatis_MC2_155 NRLDDVIIFDGLNPEELVQIVDIQLAQLAKRLEQRRLTLEVSLPAKKWLG
TH_2647|M.thermoresistible__bu NRLDDIIIFDGLNPEELVSIVDIQLAQLAKRLEQRRLTLQVSEPAKQWLA
MMAR_0645|M.marinum_M NRLDDVLIFDALNPDELVQIVDIQLQQLDKRLAQRRLQLEVSLPAKEWLA
MUL_0127|M.ulcerans_Agy99 NRLDDVLIFDALNPDELVQIVDIQLQQLDKRLAQRRLQLEVSLPAKEWLA
Mb0391c|M.bovis_AF2122/97 NRLDDVLIFEGLNPEELVRIVDIQLAQLGKRLAQRRLQLQVSLPAKRWLA
Rv0384c|M.tuberculosis_H37Rv NRLDDVLIFEGLNPEELVRIVDIQLAQLGKRLAQRRLQLQVSLPAKRWLA
MLBr_02490|M.leprae_Br4923 NRLDDVLIFDGLNPEELVRIVDIQLEQLGKRLAQRRLQLEVSLPAKRWLA
MAV_4793|M.avium_104 NRLDDVIIFHGLEPGELVQIVDIQLAQLQKRLAQRRLTLEVSLPAKQWLA
MAB_4265c|M.abscessus_ATCC_199 NRLDDVLIFDGLNPEELVQIVDIQLGQLQKRLAQRRLTLEVSAPAKKWLA
*****:::*..*:* :** ****** ** *** **** *:** ***.**.
Mflv_0246|M.gilvum_PYR-GCK ERGFDPLYGARPLRRLVQQAIGDQLARMLLSGQVHDGDVVPVNLSADGDS
Mvan_0659|M.vanbaalenii_PYR-1 ERGFDPLYGARPLRRLIQQAIGDQLAKLLLAGDVHDGDVVPVNVSPDGDS
MSMEG_0732|M.smegmatis_MC2_155 QRGFDPLYGARPLRRLVQQAIGDQLAKMLLAGEVHDGDIVPVNVSPDGES
TH_2647|M.thermoresistible__bu QRGFDPQYGARPLRRLIQKVIGDQLAKLLLAGEVHDGDVVPVNVAPDGNS
MMAR_0645|M.marinum_M QRGFDPVYGARPLRRLVQQAIGDQLAKMLLAGQVHDGDTVPVNVSADGES
MUL_0127|M.ulcerans_Agy99 QRGFDPVYGARPLRRLVQQAIGDQLAKMLLAGQVHDGDTVPVNVSADGES
Mb0391c|M.bovis_AF2122/97 QRGFDPVYGARPLRRLVQQAIGDQLAKMLLAGQVHDGDTVPVNVSPDADS
Rv0384c|M.tuberculosis_H37Rv QRGFDPVYGARPLRRLVQQAIGDQLAKMLLAGQVHDGDTVPVNVSPDADS
MLBr_02490|M.leprae_Br4923 QHGFDPVYGARPLRRLVQQAIGDQLAKMLLAGEVHDGDTLPVNVRPDGEA
MAV_4793|M.avium_104 HRGFDPVYGARPLRRLVQQAIGDQLAKQLLAGQVHDGDTVPVNVSPDGDS
MAB_4265c|M.abscessus_ATCC_199 ARGFDPIYGARPLRRLVQQSIGDQLAKQLLAGEVHDGDVVPVNVSADGES
:**** *********:*: ******: **:*:***** :***: .*.::
Mflv_0246|M.gilvum_PYR-GCK LVLG
Mvan_0659|M.vanbaalenii_PYR-1 LVLG
MSMEG_0732|M.smegmatis_MC2_155 LVLG
TH_2647|M.thermoresistible__bu LVLG
MMAR_0645|M.marinum_M LVLG
MUL_0127|M.ulcerans_Agy99 LVFG
Mb0391c|M.bovis_AF2122/97 LILG
Rv0384c|M.tuberculosis_H37Rv LILG
MLBr_02490|M.leprae_Br4923 LILG
MAV_4793|M.avium_104 LILG
MAB_4265c|M.abscessus_ATCC_199 LILG
*::*