For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEISETSGTSETDPIDPPVPIPDVPGGEAGAEGLPPLSALSLRQRLVVESSAVGDIALRTYASSLLSTTV APAVVAHALRRLDSGGERANLNFYAELAAEQDPRKSFPAPTELPRISARPASPLAEWIARGSVDDISFTS GFRAINPNMRQHWSALTSNNVVRAQHWRHDDGPRPTLCVIHGFMGSSYLLNGLFFSLPWYYRSGYDVLLY TLPFHGKRAERFSPFSGFGFFASGLSGFAESMAQAVYDFRSIVDYLHHTGVTRIALTGISLGGYTSALVS SVENRLEAVIPNCPVVTPAKLFEQWFPASKLVRLGLCLSSISRGELDAGLAFHSPLNYRPQLPKHRRMII AGLGDRMAPPDHAETLWEHWDRCALHWFPGSHVLHVSQLDYLRRMTAFLGDFMFD*
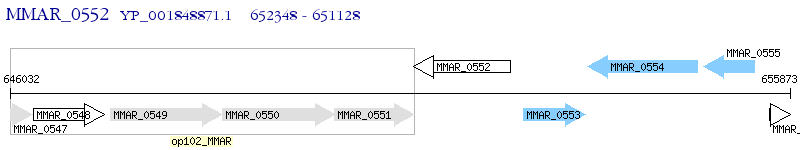
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0552 | - | - | 100% (406) | hypothetical protein MMAR_0552 |
| M. marinum M | MMAR_3425 | - | 6e-24 | 31.33% (300) | hypothetical protein MMAR_3425 |
| M. marinum M | MMAR_2072 | - | 4e-23 | 28.66% (321) | hypothetical protein MMAR_2072 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0301c | - | 0.0 | 79.20% (399) | hypothetical protein Mb0301c |
| M. gilvum PYR-GCK | Mflv_0317 | - | 2e-69 | 62.89% (194) | hypothetical protein Mflv_0317 |
| M. tuberculosis H37Rv | Rv0293c | - | 0.0 | 79.20% (399) | hypothetical protein Rv0293c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4327 | - | 1e-122 | 55.74% (366) | hypothetical protein MAB_4327 |
| M. avium 104 | MAV_4861 | - | 0.0 | 75.25% (396) | hypothetical protein MAV_4861 |
| M. smegmatis MC2 155 | MSMEG_0625 | - | 1e-151 | 62.12% (396) | hypothetical protein MSMEG_0625 |
| M. thermoresistible (build 8) | TH_1308 | - | 1e-155 | 64.39% (396) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1215 | - | 0.0 | 99.26% (406) | hypothetical protein MUL_1215 |
| M. vanbaalenii PYR-1 | Mvan_0423 | - | 1e-158 | 64.99% (397) | hypothetical protein Mvan_0423 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0552|M.marinum_M MSEISETSGTSETDPIDPPVPIPDVPGGEA--GAEGLPPLSALSLRQRLV
MUL_1215|M.ulcerans_Agy99 MSEISETSGTSETDPIDPPVPIPDVPGGEA--GAEGLPPLSALSLRQRLV
Mb0301c|M.bovis_AF2122/97 ------MSGTFTADAIGPPVPIPDVPGADA--GAEGLPSRSVLSARQRIL
Rv0293c|M.tuberculosis_H37Rv ------MSGTFTADAIGPPVPIPDVPGADA--GAEGLPSRSVLSARQRIL
MAV_4861|M.avium_104 --------MAPDLDPFDPPIPVPDVPGADVPPGAGGLPPTSALSPRQRMV
MSMEG_0625|M.smegmatis_MC2_155 -------MEPSSVDPVDPPIPIPDVPGADA--TSRGLPRRIDLTLRQKAI
TH_1308|M.thermoresistible__bu ------VGDSQSTAPVDPSFPVPDVPGADA--GAGGLPRRADLTLAQRIT
Mflv_0317|M.gilvum_PYR-GCK ----MRDVEPDSVPDEVLPLPVPDVRGADA--TARGLPRRVDLTLRQRLI
Mvan_0423|M.vanbaalenii_PYR-1 ----MTESHAPVDP----PIPVPDVPGADA--TARGLPRRQDLALRQRLI
MAB_4327|M.abscessus_ATCC_1997 ------MTAELSEHIEQPRITPRPAPTARP-----QLPRPGDLTLRERAM
.. . . ** *: ::
MMAR_0552|M.marinum_M VESSAVGDIALRTYASSLLSTTVAPAVVAHALRRLDSGGERANLNFYAEL
MUL_1215|M.ulcerans_Agy99 VESSAVGDIALRTYASSLLSTTVAPAVVAHALRRLDSGGERANLNFYAEL
Mb0301c|M.bovis_AF2122/97 VESSAIADVALRTAVASVLSATVTPAVVANALRHVNEGSERSNLNFYAEL
Rv0293c|M.tuberculosis_H37Rv VESSAIADVALRTAVASVLSATVTPAVVANALRHVNEGSERSNLNFYAEL
MAV_4861|M.avium_104 VEASALGDLALRTWVASLLSTTVAPFVVANALRHADAATERANLEFYAEL
MSMEG_0625|M.smegmatis_MC2_155 VDASAAADIALRTGVASVVAASMLPRTAATILRRRDSRIEHEHMRFYADL
TH_1308|M.thermoresistible__bu VDASLAADLALRTAVASLIGTTMLPPFVGALLNRDASRAERRHLRFYAEL
Mflv_0317|M.gilvum_PYR-GCK VDSSAVADLALRTSIASLLSVTMVP-TLAQALNRSAAATEQEHLRFYAEL
Mvan_0423|M.vanbaalenii_PYR-1 VDSSAVADLALRTSIASLLSATMVP-SVSKALRRSEAAAEQDHLRFYAEL
MAB_4327|M.abscessus_ATCC_1997 VETSALTDIALRTVGALSVMGVVAP---SLVIDREFVARERDNLAFYGEL
*::* *:**** : : : * . : : *: :: **.:*
MMAR_0552|M.marinum_M AAEQDPRKSFPAPTELPRISARPASPLAEWIARGSVDDISFTSGFRAINP
MUL_1215|M.ulcerans_Agy99 AAEQDPRKSFPAPTELPRISARPASPLAEWIARGSVDDISFTSGFRAKNP
Mb0301c|M.bovis_AF2122/97 AAAHDPAKSFPAPTELPKVTSRPASPLTEWVARGTVDNIAFASGFRAINP
Rv0293c|M.tuberculosis_H37Rv AAAHDPAKSFPAPTELPKVTSRPASPLTEWVARGTVDNIAFASGFRAINP
MAV_4861|M.avium_104 GAAADPDRSFPPPTEPPRVTSRRASPLAEWVARGTVDNLAFDSSFTAVNP
MSMEG_0625|M.smegmatis_MC2_155 AEAKDPERSFPRPTEPPRVSSRPANPVAEWVAHGRVENIRFQSSFRAVNP
TH_1308|M.thermoresistible__bu AEQRDPARAFPAPTTVPRVTTRPANPVAEFVARGHVHNIRFDSDFEAINP
Mflv_0317|M.gilvum_PYR-GCK AAAQDPQLSFPAPQSPPRVSSRAANPVAEWVAHGKVRNIRFESGFRAVNP
Mvan_0423|M.vanbaalenii_PYR-1 AAARDPQVSFPAPQSPPRVSSRPANPVAEWIARGNVRNIRFDSSFRAVNP
MAB_4327|M.abscessus_ATCC_1997 AAMGDADLSFPEPEITPVVFSRPAGRVAQALAKATVENLDFESPYQTRNP
. *. :** * * : :* *. ::: :*:. * :: * * : : **
MMAR_0552|M.marinum_M NMRQHWSALTSNNVVRAQHWRHDDGPRPTLCVIHGFMGSSYLLNGLFFSL
MUL_1215|M.ulcerans_Agy99 NMRQHWSALTSNNVVRAQHWRHDDGPRPTLCVIHGFMGSSYLLNGLFFSL
Mb0301c|M.bovis_AF2122/97 TMRQRWSALTANNIVHAQHWRHRDGPRPTLCVIHGFMGSSYLLNGLFFSL
Rv0293c|M.tuberculosis_H37Rv TMRQRWSALTANNIVHAQHWRHRDGPRPTLCVIHGFMGSSYLLNGLFFSL
MAV_4861|M.avium_104 AMRARWSGWGRNNVVRAQHWRHDDGPRPTLCVIHGFLGSSYLANGRFFSL
MSMEG_0625|M.smegmatis_MC2_155 ALRERCRGHVRNNIVRAQHWRHNDGPHPTLCVIHGFMGSPYLANGLFLSL
TH_1308|M.thermoresistible__bu ELRDRCRGYRRNNVVRAQHWRHSDGPRPTLCVIHGFMGSAYLANGLFLSL
Mflv_0317|M.gilvum_PYR-GCK ALRDHRAGFARNNIVHAQHWRHDDGPRPTLCLIHGFMGSAYLFNGLFFSL
Mvan_0423|M.vanbaalenii_PYR-1 ALRDQCAGFVRNNVVHAQHWRHDDGPRPTLCLIHGFMGSAYLFNGLFFSL
MAB_4327|M.abscessus_ATCC_1997 ALRDYWKRFPHNSVARAQHWRHDDGPRPTLCVIHGFFGSPYLLNGAFFAL
:* *.:.:****** ***:****:****:**.** ** *::*
MMAR_0552|M.marinum_M PWYYRSGYDVLLYTLPFHGKRAERFSPFSGFGFFASGLSGFAESMAQAVY
MUL_1215|M.ulcerans_Agy99 PWYYRSGYDVLLYTLPFHGKRAERFSPFSGFGFFASGLSGFAESMAQAVY
Mb0301c|M.bovis_AF2122/97 PWYYRSGYDVLLYTLPFHGQRAEKFSPFSGFGYFTSGLSGFAEAMAQAVY
Rv0293c|M.tuberculosis_H37Rv PWYYRSGYDVLLYTLPFHGQRAEKFSPFSGFGYFTSGLSGFAEAMAQAVY
MAV_4861|M.avium_104 PWYYRAGYDVLMYTLPFHGKRAERFSPFSGFGYFAGGLSGFAEAMAQAVH
MSMEG_0625|M.smegmatis_MC2_155 PWFYRAGYDVLLYTLPFHGLRAERFSPFSGYGYFANGLAGFAEAMAQAVH
TH_1308|M.thermoresistible__bu PWFYRSGYDVLLYTLPFHGRRAERNSPFSGYGFFAHGLAGFAEAMAQAVH
Mflv_0317|M.gilvum_PYR-GCK PWFYRCGYDVVLYTR--QGPSADHRRPR----------------------
Mvan_0423|M.vanbaalenii_PYR-1 PWFYRCGYDVMLYTLPFHGRRAEKYSPFSGHGYFAHGMAGFAEAMAQAVH
MAB_4327|M.abscessus_ATCC_1997 PWFFRNGYDVLLYTLPFHGRRAEKYSPFSGAGYFSHGFTGFSESMCQAVH
**::* ****::** :* *:: *
MMAR_0552|M.marinum_M DFRSIVDYLHHTGVTRIALTGISLGGYTSALVSSVENRLEAVIPNCPVVT
MUL_1215|M.ulcerans_Agy99 DFRSIVDYLHHTGVTRIALTGISLGGYTSALVSSVENRLEAVIPNCPVVT
Mb0301c|M.bovis_AF2122/97 DFRSIVDYLRHIGVDRIALTGISLGGYTSALLASVESRLEAVIPNCPVVM
Rv0293c|M.tuberculosis_H37Rv DFRSIVDYLRHIGVDRIALTGISLGGYTSALLASVESRLEAVIPNCPVVM
MAV_4861|M.avium_104 DFRSLIDYLRHTGVERIALTGISLGGYTSALVASVDDRLEAVIPNCPVVT
MSMEG_0625|M.smegmatis_MC2_155 DFRSVMDHLEFTGVDRIALTGISLGGFTSALLASVDHRIQAVIPNVPVVT
TH_1308|M.thermoresistible__bu DFRSLMNYLEFTGVDRIALTGISLGGYTSALLAAVDDRIQAVIPNVPVVT
Mflv_0317|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0423|M.vanbaalenii_PYR-1 DFRSIIDYLEFTGVDRIALTGMSLGGYTSALIASVDDRVQAVIPNVPVVT
MAB_4327|M.abscessus_ATCC_1997 DFRIFVNYLRGTGVEKIGLTGLSLGGYTTGLISTVERRIDVTIPNVPVVD
MMAR_0552|M.marinum_M PAKLFEQWFPASKLVRLGLCLSSISRGELDAGLAFHSPLNYRPQLPKHRR
MUL_1215|M.ulcerans_Agy99 PAELFEQWFPASKLVRLGLCLSSISRGELDAGLAFHSPLNYRPQLPKHRR
Mb0301c|M.bovis_AF2122/97 PAKLFDEWFPANKLVKLGLRLTNISRDELIAGLAYHGPLNYRPLLPKDRR
Rv0293c|M.tuberculosis_H37Rv PAKLFDEWFPANKLVKLGLRLTNISRDELIAGLAYHGPLNYRPLLPKDRR
MAV_4861|M.avium_104 PATLFDEWFPANKLVGLGLRSSDISRAQLAAGLAYHCPLTYRPLLAKDRR
MSMEG_0625|M.smegmatis_MC2_155 PDRTVDEWFPANMLVALERHIAHTDPELTEAAARYASPLHYQPLVAKERR
TH_1308|M.thermoresistible__bu PQAAFDDWFPANKLVALTNAVTAIDRELAGAAGMYHSPLNYRPLVPKERR
Mflv_0317|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0423|M.vanbaalenii_PYR-1 PDRTVDEWFPANKVVALRDWIAGTDTELVGAATMYSSPLNYRPLIPKDRR
MAB_4327|M.abscessus_ATCC_1997 VKSLLHSWIPAEPTIKLAARLGHLDQGRLDGALSYSSPLTYQPVVPREQR
MMAR_0552|M.marinum_M MIIAGLGDRMAPPDHAETLWEHWDRCALHWFPGSHVLHVSQLDYLRRMTA
MUL_1215|M.ulcerans_Agy99 MIIAGLGDRMAPPDHAEPLWEHWDRCALHWFPGSHVLHVSQLDYLRRMTA
Mb0301c|M.bovis_AF2122/97 MIITGLGDRMAPPEHAVTLWKQWDRCALHWFPGSHLLHVSQLDYLRRMTV
Rv0293c|M.tuberculosis_H37Rv MIITGLGDRMAPPEHAVTLWKQWDRCALHWFPGSHLLHVSQLDYLRRMTV
MAV_4861|M.avium_104 MIITGLGDRMAPPEHAVKLWQHWDHCALHWFPGSHVLHISQLDYLRRMTA
MSMEG_0625|M.smegmatis_MC2_155 LIITGLGDRLAPPQQAELLWEHWDRCAFHWFPGNHLLHVSQPDYLRRMTR
TH_1308|M.thermoresistible__bu LIITGLGDRFAPPQQAELLWKHWDRCAFHWFPGNHLLHVSQPDYLRRMTR
Mflv_0317|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0423|M.vanbaalenii_PYR-1 LIIAGLGDRLAPPEQAEILWRHWDHCAFHWFPGNHVLHVSQPDYLRRMTR
MAB_4327|M.abscessus_ATCC_1997 LIITGLGDRLAPPEQSEMLWEHWDRCALHWFPGNHILHVNQGDYLRRMTE
MMAR_0552|M.marinum_M FLGDFMFD
MUL_1215|M.ulcerans_Agy99 FLGDFMFD
Mb0301c|M.bovis_AF2122/97 FLQGLMFD
Rv0293c|M.tuberculosis_H37Rv FLQGLMFD
MAV_4861|M.avium_104 FLQQVMF-
MSMEG_0625|M.smegmatis_MC2_155 FLRGFMFD
TH_1308|M.thermoresistible__bu FLGDVMFG
Mflv_0317|M.gilvum_PYR-GCK --------
Mvan_0423|M.vanbaalenii_PYR-1 FMGAFMFD
MAB_4327|M.abscessus_ATCC_1997 FLDRHLNV