For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAATGPTTMSMTSMMSRVKWMLRARPADYLLAMSVAGASLPVVGKHLEPLGCVTAMGVWGARHTPEFFAA RAKDLFTPGINVTRQRDRDSTNEVSAAALRGIVSTADLDVEWPAPQRTPPVCEAFRHRRYLHRGGVHYGN SPAQVLDVWRRDDLPTEPAPVLIFVPGGAWIHGSRAIQGYALLSRLAEQGWVCLSIDYRVAPHHRWPRHI HDVKAAIAWARANVDKFGGDRNFIAVAGCSAGGHLSALAGLTANDPDYQSELPEGSDTSVDAAVGIYGRY DWEDRSTAERARFVDFLERVVVKRSIKRHPQVFRDASPVARVHTNAPPFLVIHGSRDGVIPVAQARSFVE RLRAVSRSLVAYVELPGAGHGFDLLDGARTGSATHAISLFLNQIHRTRAQLSKEVI
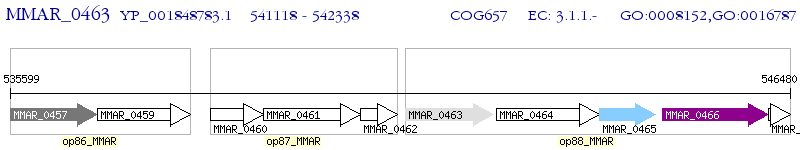
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0463 | lipC | - | 100% (406) | esterase LipC |
| M. marinum M | MMAR_2232 | lipO | 1e-76 | 50.53% (285) | esterase LipO |
| M. marinum M | MMAR_3429 | lipM | 8e-64 | 45.45% (286) | membrane-bound esterase LipM |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0226 | lipC | 0.0 | 82.52% (389) | esterase LipC |
| M. gilvum PYR-GCK | Mflv_0440 | - | 1e-165 | 68.84% (398) | alpha/beta hydrolase domain-containing protein |
| M. tuberculosis H37Rv | Rv0220 | lipC | 0.0 | 82.52% (389) | esterase LipC |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3447c | - | 1e-13 | 32.02% (253) | putative lipase/esterase |
| M. avium 104 | MAV_4953 | - | 1e-175 | 72.84% (394) | alpha/beta hydrolase fold |
| M. smegmatis MC2 155 | MSMEG_0289 | - | 1e-154 | 67.95% (365) | alpha/beta hydrolase fold family protein |
| M. thermoresistible (build 8) | TH_2755 | - | 1e-156 | 65.82% (395) | PUTATIVE Alpha/beta hydrolase fold-3 domain protein |
| M. ulcerans Agy99 | MUL_1112 | lipC | 0.0 | 99.74% (392) | esterase LipC |
| M. vanbaalenii PYR-1 | Mvan_0227 | - | 1e-169 | 69.87% (395) | alpha/beta hydrolase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0440|M.gilvum_PYR-GCK MTERPDTRYPGPTLTTRLEWLLNAGPSDYMLALSVASASLPVIGKHLEPL
Mvan_0227|M.vanbaalenii_PYR-1 MTERPDIRYPGPTLTTRLQWLLNAGPSDFMLAMSVASASLPVIGKHLEPL
MSMEG_0289|M.smegmatis_MC2_155 -----------------------------------------MIGKHFEPL
TH_2755|M.thermoresistible__bu MTVRPDG---GSSAWLCARRLLRAGPADHLLALSAASGSLPIVGRHLEPL
MMAR_0463|M.marinum_M MAATGPTTMSMTSMMSRVKWMLRARPADYLLAMSVAGASLPVVGKHLEPL
MUL_1112|M.ulcerans_Agy99 --------------MSRVKWMLRARPADYLLAMSVAGASLPVVGKHLEPL
Mb0226|M.bovis_AF2122/97 ---MNQRRAAGSTGVAYIRWLLRARPADYMLALSVAGGSLPVVGKHLKPL
Rv0220|M.tuberculosis_H37Rv ---MNQRRAAGSTGVAYIRWLLRARPADYMLALSVAGGSLPVVGKHLKPL
MAV_4953|M.avium_104 ---------MAPGMTPYLKWLLRARPADYALAVSLAGASLPVVGRHLEPL
MAB_3447c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0440|M.gilvum_PYR-GCK GALTAAGVWGARHAPDFVGALAKSLVAPNDVGKKEKQQQERDSTNAITQA
Mvan_0227|M.vanbaalenii_PYR-1 GALTAAGIWGTRHAPDFVGALAKSLITPNDAERK---KQERDCTNAVTEA
MSMEG_0289|M.smegmatis_MC2_155 GSMTAMSIWGLRHLPDFVAASAKSRLAPGAAAMR---REEREQTTAAAVA
TH_2755|M.thermoresistible__bu GSAAAMSLWGFRHLPDFVGATVKSWLAPGSAELR---RRQREMTNEVSRQ
MMAR_0463|M.marinum_M GCVTAMGVWGARHTPEFFAARAKDLFTPGINVTR---QRDRDSTNEVSAA
MUL_1112|M.ulcerans_Agy99 GCVTAMGVWGARHTPEFVAARAKDLFTPGINVTR---QRDRDSTNEVSAA
Mb0226|M.bovis_AF2122/97 GGVTAIGVWGARHASDFLSATAKDLLTPGINEVR---RRDRASTQEVSVA
Rv0220|M.tuberculosis_H37Rv GGVTAIGVWGARHASDFLSATAKDLLTPGINEVR---RRDRASTQEVSVA
MAV_4953|M.avium_104 AGLTAMGMWGARHAPEVISAATRDMFAPGGGDAR---RRDRESTQQVSLA
MAB_3447c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0440|M.gilvum_PYR-GCK ALRGLVSQK--------------DLEIEWPAAERTPPVWKMREHRRHVHR
Mvan_0227|M.vanbaalenii_PYR-1 ALRDVVGPK--------------DLEIEWPAAERTPPVWRMREHRRHVHR
MSMEG_0289|M.smegmatis_MC2_155 ALRNVVPPA--------------ELAAEWPAPQNLPPLWNVLEHRRSVYR
TH_2755|M.thermoresistible__bu ALRGLLPADPAAPGASEVSSGSAGSALDWPAPDRVPPLWRMLQHRRHLYR
MMAR_0463|M.marinum_M ALRGIVSTA--------------DLDVEWPAPQRTPPVCEAFRHRRYLHR
MUL_1112|M.ulcerans_Agy99 ALRGIVSTA--------------DLDVEWPAPQRTPPVCEAFRHRRYLHR
Mb0226|M.bovis_AF2122/97 ALRGIVSPD--------------DLAVEWPAPERTPPVCGALRHRRYVHR
Rv0220|M.tuberculosis_H37Rv ALRGIVSPD--------------DLAVEWPAPERTPPVCGALRHRRYVHR
MAV_4953|M.avium_104 ALRGLVSAA--------------ELESQWPVAQRTPPIWDGVRRHRHLYR
MAB_3447c|M.abscessus_ATCC_199 --MNLIQEG-----------------------EAMP------QHRR----
.:: : * .::*
Mflv_0440|M.gilvum_PYR-GCK TSVRYGPRPTQLLDVWRRDDVPAD-APVMIFVPGGAWVHGGRMLQGYALM
Mvan_0227|M.vanbaalenii_PYR-1 TSVRYGPRPTQLLDVWRRDDLPAEPAPVLIFVPGGAWVHGSRMLQGYALM
MSMEG_0289|M.smegmatis_MC2_155 TSVRYGDDPAQLLDVWRPKELPAQPAPVLLFVPGGAWVHGSRVLQGYALM
TH_2755|M.thermoresistible__bu GAVRYGDHTENVLDVWRRKDLPAQPAPVLIFVPGGAWVHGSRMLQGYALL
MMAR_0463|M.marinum_M GGVHYGNSPAQVLDVWRRDDLPTEPAPVLIFVPGGAWIHGSRAIQGYALL
MUL_1112|M.ulcerans_Agy99 GGVHYGNSPAQVLDVWRRDDLPTEPAPVLIFVPGGAWIHGSRAIQGYALL
Mb0226|M.bovis_AF2122/97 RRVLYGDDPAQLLDVWRRKDMPTKPAPVLIFVPGGAWVHGSRAIQGYAVL
Rv0220|M.tuberculosis_H37Rv RRVLYGDDPAQLLDVWRRKDMPTKPAPVLIFVPGGAWVHGSRAIQGYAVL
MAV_4953|M.avium_104 RAVHYGHHPAQVLDVWRRKDLPAQPAPVLIFVPGGAWVHGKSMGQGSALM
MAB_3447c|M.abscessus_ATCC_199 --LTYGDHPSQYVDILEPD---EPAAALVHVIHGGFWREALSAELTAPIA
: ** . : :*: . . *.:: .: ** * .. .:
Mflv_0440|M.gilvum_PYR-GCK HHLAEMGWVCLSVEYR-VAPHNPWPAHINDVKTAIAWARANVDKFGGNRD
Mvan_0227|M.vanbaalenii_PYR-1 SHLAERGWVCLSIDYR-VAPHNPWPAHVADVKTAIAWARANVDKFGGDRN
MSMEG_0289|M.smegmatis_MC2_155 SHLAQQGWVCLSVDYR-VSPHHRWPRHIHDVKAAIAWARANVDKFGGDRH
TH_2755|M.thermoresistible__bu SHLAEQGWVCLSIDYR-VAPQHPWPAHLTDVKTAIAWARANVDKFGGDRD
MMAR_0463|M.marinum_M SRLAEQGWVCLSIDYR-VAPHHRWPRHIHDVKAAIAWARANVDKFGGDRN
MUL_1112|M.ulcerans_Agy99 SRLAEQGWVCLSIDYR-VAPHHRWPRHIHDVKAAIAWARANVDKFGGDRN
Mb0226|M.bovis_AF2122/97 SRLAAQGWVCLSIDYR-VAPHHRWPRHILDVKTAIAWARANVDKFGGDRN
Rv0220|M.tuberculosis_H37Rv SRLAAQGWVCLSIDYR-VAPHHRWPRHILDVKTAIAWARANVDKFGGDRN
MAV_4953|M.avium_104 SRLAEQGWVCLAIDYR-VAPHHRWPRHIVDVKTAIAWARANVDKFGGDRN
MAB_3447c|M.abscessus_ATCC_199 RDLCTDGFLVANIEYRRVGGGGGWPETFEDVKAAIAAVRQAEPDLVRS--
*. *:: ::** *. ** . ***:*** .* .: .
Mflv_0440|M.gilvum_PYR-GCK FVAIAGTSAGGHLAALAGLTANDPEMQGELPEGSDTSVDAVVGIYGRYDW
Mvan_0227|M.vanbaalenii_PYR-1 FVAISGASAGGHLAALAGLTANDPEMQDELPEGSDTSVDAVVGIYGRYDW
MSMEG_0289|M.smegmatis_MC2_155 FIAIAGCSAGGHLAALAGLTPNHPELQGDLPDGADTSVDAVVGIYGRYDW
TH_2755|M.thermoresistible__bu FVAVAGCSAGGHLAALAGLTENDPDMQTELPEGSDTSVDAVVGVYGRYDW
MMAR_0463|M.marinum_M FIAVAGCSAGGHLSALAGLTANDPDYQSELPEGSDTSVDAAVGIYGRYDW
MUL_1112|M.ulcerans_Agy99 FIAVAGCSAGGHLSALAGLTANDPDYQSELPEGSDTSVDAAVGIYGRYDW
Mb0226|M.bovis_AF2122/97 FIAVAGCSAGGHLSALAGLTANDPQYQAELPEGSDTSVDAVVGIYGRYDW
Rv0220|M.tuberculosis_H37Rv FIAVAGCSAGGHLSALAGLTANDPQYQAELPEGSDTSVDAVVGIYGRYDW
MAV_4953|M.avium_104 FVAIAGCSAGGHLSALAGLTPDDPQYRGMLPEGADTSVDAVVGIYGRYDW
MAB_3447c|M.abscessus_ATCC_199 --LAVGHSAGGHLSLLA-LAAGLADFAVALAPVSDVDRALRENLGG----
* ******: ** *: . .: *. :*.. .: *
Mflv_0440|M.gilvum_PYR-GCK EDKSTVERVRFMDFLERVVVKRKFDKHPDVFRKASPMARIHSEAPPFLVI
Mvan_0227|M.vanbaalenii_PYR-1 EDKSTVERVRFMDFLERVVVRRKFDRHPELFRKASPMARVHPEAPPFLVV
MSMEG_0289|M.smegmatis_MC2_155 EDRSTVERERFVDFLERVVVKRSIERHPEIFRLASPIAQVHADAPPFLVV
TH_2755|M.thermoresistible__bu VDRSTAERVRFLDFLERVVVRKRLDRHPEVFHRASPMHRVHGDAPPFLLV
MMAR_0463|M.marinum_M EDRSTAERARFVDFLERVVVKRSIKRHPQVFRDASPVARVHTNAPPFLVI
MUL_1112|M.ulcerans_Agy99 EDRSTAERARFVDFLERVVVKRSIKRHPQVFRDASPVARVHTNAPPFLVI
Mb0226|M.bovis_AF2122/97 EDRSTPERARFVDFLERVVVQRTIDRHPEVFRDASPIQRVTRNAPPFLVI
Rv0220|M.tuberculosis_H37Rv EDRSTPERARFVDFLERVVVQRTIDRHPEVFRDASPIQRVTRNAPPFLVI
MAV_4953|M.avium_104 EDRSTPERVRFVDFLERVVVRKSIARHPEVFRDASPIARVHRNAPPFLVI
MAB_3447c|M.abscessus_ATCC_199 -----GAAAEFLG---------VAGSSPREVRTASPIGNLG-HRRQHVLI
.*:. * .: ***: .: . .:::
Mflv_0440|M.gilvum_PYR-GCK HGTGDSVIPVAQAQSFVERLRNVSRSVVGYVELPGAGHGFDMTDGARTGA
Mvan_0227|M.vanbaalenii_PYR-1 HGTGDSVIPVAQAQSFVERLRGVSRSVVGYVELPGAGHGFDMTDGARTGA
MSMEG_0289|M.smegmatis_MC2_155 HGSADTVIPVAQARSFVDRLGAASRAPVSYLELPGAGHGFDMTDGARTGA
TH_2755|M.thermoresistible__bu HGSADTVIPVRQARDFADRLRSVSRSAVSYLELPGAGHGFDLLDGARTGS
MMAR_0463|M.marinum_M HGSRDGVIPVAQARSFVERLRAVSRSLVAYVELPGAGHGFDLLDGARTGS
MUL_1112|M.ulcerans_Agy99 HGSRDGVIPVAQARSFVERLRAVSRSLVAYVELPGAGHGFDLLDGARTGS
Mb0226|M.bovis_AF2122/97 HGSRDCVIPVEQARSFVERLRAVSRSQVGYLELPGAGHGFDLLDGARTGP
Rv0220|M.tuberculosis_H37Rv HGSRDCVIPVEQARSFVERLRAVSRSQVGYLELPGAGHGFDLLDGARTGP
MAV_4953|M.avium_104 HGSKDSVIPVEQARSFVERLRAVSHSTVGYLELPGAGHGYDLIDGERAGA
MAB_3447c|M.abscessus_ATCC_199 HGLRDVNVPVEHSQHYVSAARASG-TPVDYFEFANLDH-FDLIDPSSSAW
** * :** ::: :.. . : * *.*:.. .* :*: * :.
Mflv_0440|M.gilvum_PYR-GCK AATAIGLFLNHIHRNRSLVGAKEVI
Mvan_0227|M.vanbaalenii_PYR-1 ASTAIGLFLNHIHRNRTLIGAKEVI
MSMEG_0289|M.smegmatis_MC2_155 MATAIGLFLKEIHRNRTRNTAKAVI
TH_2755|M.thermoresistible__bu AATAVGLFLDHVQRNRSRVRGRAVG
MMAR_0463|M.marinum_M ATHAISLFLNQIHRTRAQL-SKEVI
MUL_1112|M.ulcerans_Agy99 ATHAISLFLNQIHRTRAQL-SKEVI
Mb0226|M.bovis_AF2122/97 TAHAIALFLNQVHRSRAQF-AKEVI
Rv0220|M.tuberculosis_H37Rv TAHAIALFLNQVHRSRAQF-AKEVI
MAV_4953|M.avium_104 AAHVCSLFLDQVHRSKTRIGAKEVI
MAB_3447c|M.abscessus_ATCC_199 RVAKQWIRYQLI-------------
: . :