For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DNQRADSQLDEDEAMLVATVRAFIDRDVKPSVREVEHANTYPQAWIEQMKRIGIYGLAIGEAYGGSPVSM PCYVQVTQELARGWMSLAGAMGGHTVVAKLLMLFGTDEQKSTYLPAMATGELRATMALTEPGGGSDLQNM STTAMADGPDQLLITGAKTWISNARRSGLIALLCKTDPHATPRHHGISIVLVEPGPGLNISADLPKLGYK GVEACELSFDNYRAPTSAILGGQPGQGFSQMMKGLETGRIQVAARALGVASAALDDALAYAQQRHSFGQP IWKHQAIGNYLADMATKLTAARQLTHYAAQRYDSGQRCDMEAGMAKLFASETAMEIALNAVRIHGGYGYS TEYDVERYFRDAPLMIVGEGTNEIQRNVIARQLIARGGI*
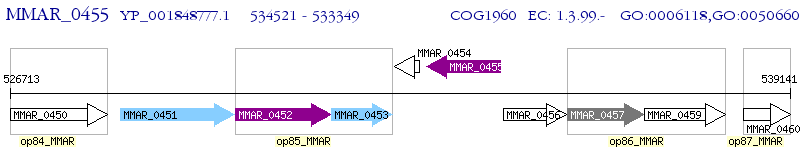
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0455 | fadE3_2 | - | 100% (390) | acyl-CoA dehydrogenase FadE3_2 |
| M. marinum M | MMAR_4161 | fadE3 | 0.0 | 98.46% (390) | acyl-CoA dehydrogenase FadE3 |
| M. marinum M | MMAR_4030 | - | 1e-99 | 47.78% (383) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0221c | fadE3 | 0.0 | 86.79% (386) | acyl-CoA dehydrogenase FADE3 |
| M. gilvum PYR-GCK | Mflv_0446 | - | 0.0 | 83.25% (382) | butyryl-CoA dehydrogenase |
| M. tuberculosis H37Rv | Rv0215c | fadE3 | 1e-178 | 86.44% (354) | acyl-CoA dehydrogenase FADE3 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 6e-56 | 35.36% (379) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3618c | - | 3e-59 | 36.15% (379) | acyl-CoA dehydrogenase FadE |
| M. avium 104 | MAV_4958 | - | 0.0 | 84.29% (382) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1991 | - | 1e-161 | 72.44% (381) | isovaleryl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3837 | - | 0.0 | 84.82% (382) | PUTATIVE Butyryl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_4024 | fadE3 | 0.0 | 98.46% (390) | acyl-CoA dehydrogenase FadE3 |
| M. vanbaalenii PYR-1 | Mvan_0221 | - | 1e-179 | 82.98% (382) | butyryl-CoA dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0446|M.gilvum_PYR-GCK ----------MVDGLSSEEAMLVQTVRAFIDRDVKPTVRDVEHANTYPEA
Mvan_0221|M.vanbaalenii_PYR-1 ----------MVEGLSSEEAMLVETVRTFIDREVRPTVRDVEHANTYPEA
MMAR_0455|M.marinum_M -----MDNQRADSQLDEDEAMLVATVRAFIDRDVKPSVREVEHANTYPQA
MUL_4024|M.ulcerans_Agy99 ----MMNNDSPDSQLDEDEAMLVATVRAFIDRDVRPSVREVEHANTYPQA
Mb0221c|M.bovis_AF2122/97 -----MRN-----ELNDDEAMLVATVRAFIDRDVKPTVREVEHANSYPEA
Rv0215c|M.tuberculosis_H37Rv -----MRN-----ELNDDEAMLVATVRAFIDRDVKPTVREVEHANSYPEA
MAV_4958|M.avium_104 --------------MNDEEDMLVATVRAFIDREVKPTVREVEHADAYPEA
TH_3837|M.thermoresistible__bu ----------VNVVLNDEETMLVETVRAFIDRDVKPTVREVEHANEYPQA
MSMEG_1991|M.smegmatis_MC2_155 ------------MALTAEEETIVKTVHDFVEKQVKPVVRELEHANTYPEE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
MAB_3618c|M.abscessus_ATCC_199 ---MAGNPSFDLFKLGEEHDELRAAIRGLAEKEIAPHAKDVDEKARFPQE
: :. : ::: : :::: * . :::. :*:
Mflv_0446|M.gilvum_PYR-GCK WIDQMKQIGIYGLAVPESYGGTPVSTRCYVLVTQELARGWMSLAGAMGGH
Mvan_0221|M.vanbaalenii_PYR-1 WIEKMKQIGIFGLAIPESEGGAPVSTRCYVLVTQELARGWMSLAGAMGGH
MMAR_0455|M.marinum_M WIEQMKRIGIYGLAIGEAYGGSPVSMPCYVQVTQELARGWMSLAGAMGGH
MUL_4024|M.ulcerans_Agy99 WIEQMKRIGIYGLAIGEAYGGSPVSMPCYVQVTQELARGWMSLAGAMGGH
Mb0221c|M.bovis_AF2122/97 WIEQMKHIGIYGLAIDEQYGGSPVSMPCYVQVTQELARGWMSLAGAMGGH
Rv0215c|M.tuberculosis_H37Rv WIEQMKHIGIYGLAIDEQYGGSPVSMPCYVQVTQELARGWMSLAGAMGGH
MAV_4958|M.avium_104 WIEQMKRIGIYGLAVPEEYGGSPVSMPCYVRVTEQLARGWMSLAGAMGGH
TH_3837|M.thermoresistible__bu WIEQMKRIGIYGLAIPESYGGSPVSMPCYVEVTQELSRGWMSLAGAMGGH
MSMEG_1991|M.smegmatis_MC2_155 LIETMKEIGIFGLAIPEPYGFGAVSMPCYVQVAEELARGWMSLAGAMGGH
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MAB_3618c|M.abscessus_ATCC_199 ALDALVASGFNAVHVPEEYDGQGADSVAACIVIEEVARVCASSSLIPAVN
: : *: .: : * . .. . * ::::* * : . :
Mflv_0446|M.gilvum_PYR-GCK TVVAKLLTLFGTEEQKQRYLPAMATGEIRATMALTEPGGGSDLQAMGTVA
Mvan_0221|M.vanbaalenii_PYR-1 TVVAKLLSLFGTDEQKQRYLPPMATGELRATMALTEPGGGSDLQAMSTVA
MMAR_0455|M.marinum_M TVVAKLLMLFGTDEQKSTYLPAMATGELRATMALTEPGGGSDLQNMSTTA
MUL_4024|M.ulcerans_Agy99 TVVAKLLMLFGTDEQKSTYLPAMATGELRATMALTEPGGGSDLQNMSTTA
Mb0221c|M.bovis_AF2122/97 TVVAKLLTLFGTEEQRRTYLPPMASGELRATMALTEPGGGSDLQNMSTTA
Rv0215c|M.tuberculosis_H37Rv TVVAKLLTLFGTEEQRRTYLPPMASGELRATMALTEPGGGSDLQNMSTTA
MAV_4958|M.avium_104 TVVAKLLTLFGTEDQKRAYLPRMASGEIRATMALTEPGGGSDLQNMSTTA
TH_3837|M.thermoresistible__bu TVVAKLLTLFGTEEQKQRYLPAMATGEIRATMALTEPGGGSDLQNMTTTA
MSMEG_1991|M.smegmatis_MC2_155 TVVSKLLLLFGTEEQKQKYLPRMATGELRATMALTEPGGGSDLQAMRTVA
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRA
MAB_3618c|M.abscessus_ATCC_199 KLGTMGLILNGSDELKKQVLPAIASGEAMASYALSEREAGSDAASMRTRA
.: : * * *::: : ** :*: *: **:* .*** * * *
Mflv_0446|M.gilvum_PYR-GCK RRD-GDD---LVLSGTKTWISNARRSGLIALLCKTDPDATPKHSGISVVL
Mvan_0221|M.vanbaalenii_PYR-1 RRD-GDE---LVISGAKTWISNARRSGLIALLCKTDPEATPRHSGISVVL
MMAR_0455|M.marinum_M MADGPDQ---LLITGAKTWISNARRSGLIALLCKTDPHATPRHHGISIVL
MUL_4024|M.ulcerans_Agy99 MADGPDQ---LLITGAKTWISNARRSGLIALLCKTDPHATPRHHGISIVL
Mb0221c|M.bovis_AF2122/97 LADGPEGSAGLLINGCKTWISNARRSGLFAVLCKTDPNATPRHQGMSIVL
Rv0215c|M.tuberculosis_H37Rv LADGPEGSAGLLINGCKTWISNARRSGLFAVLCKTDPNATPRHQGMSIVL
MAV_4958|M.avium_104 LPD-PDSD-GLVINGAKTWISNARRSGLIALLCKTDPKATPRHKGISILL
TH_3837|M.thermoresistible__bu LPGPGGG---LVINGAKTWISNARRSGLIALLCKTDPDATPRHRGISVVL
MSMEG_1991|M.smegmatis_MC2_155 RRD-GDD---YVINGSKTWISNARRSDLVALMCKTDPDAQPAHKGVSILL
MLBr_00737|M.leprae_Br4923 KADGDD----WILNGFKCWITNGGKSTWYTVMAVTDPDKG--ANGISAFI
MAB_3618c|M.abscessus_ATCC_199 KADGDD----WIINGSKCWITNGGKSSWYTVMAVTDPEKK--ANGISAFM
. ::.* * **:*. :* :::. ***. *:* .:
Mflv_0446|M.gilvum_PYR-GCK VDNPTPGLTVSRDLPKLGYKGVESCELAFDACRVPASAILGGTPGKGFSQ
Mvan_0221|M.vanbaalenii_PYR-1 VDNPTDGLTVSRDLPKLGYKGVESCELTFDGCRVPESAILGGAAGRGFAQ
MMAR_0455|M.marinum_M VEPGP-GLNISADLPKLGYKGVEACELSFDNYRAPTSAILGGQPGQGFSQ
MUL_4024|M.ulcerans_Agy99 VEPGP-GLNISADLPKLGYKGVEACELSFDNYRAPTSAILGGQPGQGFSQ
Mb0221c|M.bovis_AF2122/97 VEPGP-GLTVSRDLPKLGYKGVESCELSFDNLRVPVSAILGGAMGQGFSQ
Rv0215c|M.tuberculosis_H37Rv VEPGP-GLTVSRDLPKLGYKGVESCELSFDNLRVPVSAILGGAMGQGFSQ
MAV_4958|M.avium_104 VEHRP-GLTVSRDLPKLGYKGVESCELSFDNFRAPATAVLGGVAGQGFSQ
TH_3837|M.thermoresistible__bu VEQGTTGLSVSRDLPKLGYKGVESCELVFDDCRVPASAILGGEPGKGFTQ
MSMEG_1991|M.smegmatis_MC2_155 VEKVP-GFDVSRDLPKLGYKGVESCELNFTDARVPVSSLLGDDEGRGFAQ
MLBr_00737|M.leprae_Br4923 VHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIG-EPGTGFKT
MAB_3618c|M.abscessus_ATCC_199 VHKDDEGFTVGPLEHKLGIKGSPTAELYFENCRIPGDRIIG-EPGTGFKT
*. *: :. *** ** : ** * * * ::* * **
Mflv_0446|M.gilvum_PYR-GCK MMKGLETGRIQVASRALGVASAALEDALAYAQDRESFGRPIWKHQAVGHY
Mvan_0221|M.vanbaalenii_PYR-1 MMKGLETGRIQVASRALGVASAALADALAYAQDRESFGQPIWKHQAVGHY
MMAR_0455|M.marinum_M MMKGLETGRIQVAARALGVASAALDDALAYAQQRHSFGQPIWKHQAIGNY
MUL_4024|M.ulcerans_Agy99 MMKGLETGRIQVAARALGVASAALDDALAYAQQRHSFGQPIWKHQAIGNY
Mb0221c|M.bovis_AF2122/97 MMKGLETGRIQVAARALGVATAALEDSLAYAQQRESFGRPIWQHQAVGNY
Rv0215c|M.tuberculosis_H37Rv MMKGLETGRIQVAARALGVATAALEDSLAYAQQRESFGRPIWQHQAVGNY
MAV_4958|M.avium_104 MMKGLETGRIQVAARALGVATAALEDALAYAQQRESFGQPIWQHQSVGNY
TH_3837|M.thermoresistible__bu MMKGLETGRIQVAARALGVATAALEDALAYAQQRESFGQPIWKHQAIGHY
MSMEG_1991|M.smegmatis_MC2_155 MMKGLEVGRLQVAARATGVARAAFEDALRYSQERESFGKPIWQHQSVGNM
MLBr_00737|M.leprae_Br4923 ALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFM
MAB_3618c|M.abscessus_ATCC_199 ALQTLDHTRPTIGAQALGIAQGALDQAIAYTKDRKQFGKSISDFQGVQFM
: *: * :.::* *:* .*: :: *:::*..**..* .*.:
Mflv_0446|M.gilvum_PYR-GCK LADMATKLTAARQLTLYAADRYDSGER-ADMEAGMAKLFASEVAMEIALN
Mvan_0221|M.vanbaalenii_PYR-1 LADMATKLTAARQLTLYAADRYDSGAR-ADMEAGMAKLFASEVAMEIALN
MMAR_0455|M.marinum_M LADMATKLTAARQLTHYAAQRYDSGQR-CDMEAGMAKLFASETAMEIALN
MUL_4024|M.ulcerans_Agy99 LADMATKLTAARQLTHYAAQRYDSGQR-CDMEAGMAKLFASETAMEIALN
Mb0221c|M.bovis_AF2122/97 LADMATKLTAARQLTRYAAERYDSGQR-CDMEAGMAKLFASEVAMEIALN
Rv0215c|M.tuberculosis_H37Rv LADMATKLTAARQLTRYAAERYDSGQR-CDMEAGMAKLFASEVAMEIALN
MAV_4958|M.avium_104 LADMATKLTAARQLTRYAAERYDSGER-CDMEAGMAKLFASEVAMEIALN
TH_3837|M.thermoresistible__bu LADMATKLTAARQLTRYAAERYDSGQR-CDMEAGMAKLFASEVAMEIALN
MSMEG_1991|M.smegmatis_MC2_155 LADMGTKLYAARSLLLSAAEKFDAGQR-CDMEAGMAKLFASETAMQIALD
MLBr_00737|M.leprae_Br4923 LADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTD
MAB_3618c|M.abscessus_ATCC_199 LADMAMKVEAARLMVYTAAARAERGEKNLGFISSASKCFASDVAMEVTTD
****. *: *** : ** : : * .: :. :* ***: **::: :
Mflv_0446|M.gilvum_PYR-GCK AVRIHGGYGYSTEYDVERYFRDAPLMIVGEGTNEIQRNVIAGQLVARGGI
Mvan_0221|M.vanbaalenii_PYR-1 AVRIHGGYGYSTEYDVERYFRDAPLMIVGEGTNEIQRNVIAGQLVARGGI
MMAR_0455|M.marinum_M AVRIHGGYGYSTEYDVERYFRDAPLMIVGEGTNEIQRNVIARQLIARGGI
MUL_4024|M.ulcerans_Agy99 AVRIHGGYGYSTEYDVERYFRDAPLMIVGEGTNEIQRNVIARQLIARGGI
Mb0221c|M.bovis_AF2122/97 AVRIHGGYGYSTEYDVERYFRDAPLMILGEGTNEIQRNVIAGQLVARGGI
Rv0215c|M.tuberculosis_H37Rv AVRIHGGYGYSTEYDVERR-------------------------------
MAV_4958|M.avium_104 AVRIHGGYGYSQEYDVERYFRDAPLMIVGEGTNEIQRNVIARQLVTRGGI
TH_3837|M.thermoresistible__bu AVRIHGGYGYSTEFDVERYFRDAPLMIVGEGTNEIQRNVIVSQLVARNTV
MSMEG_1991|M.smegmatis_MC2_155 AVRVHGGYGYSTEYDVERYFRDAPLMIVGEGTNEIQRNVIAKQLVARGGL
MLBr_00737|M.leprae_Br4923 AVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR----
MAB_3618c|M.abscessus_ATCC_199 AVQLFGGAGYTTDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLTS---
**::.** **: :: ***
Mflv_0446|M.gilvum_PYR-GCK --
Mvan_0221|M.vanbaalenii_PYR-1 --
MMAR_0455|M.marinum_M --
MUL_4024|M.ulcerans_Agy99 --
Mb0221c|M.bovis_AF2122/97 --
Rv0215c|M.tuberculosis_H37Rv --
MAV_4958|M.avium_104 --
TH_3837|M.thermoresistible__bu --
MSMEG_1991|M.smegmatis_MC2_155 DI
MLBr_00737|M.leprae_Br4923 --
MAB_3618c|M.abscessus_ATCC_199 --