For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQQLGPKCARGQLSQPDRAWLDAARSSRSQPSTDRLSALKLSGSQLSATDARAIGQSTQIFPRHVSKISL WSTSSGCNPGLKYTFALGAMPSKKWDTGQHDRAAQRFDGMEPGPRSAAEPYTCRVNISKAIVGAATAPIR AGVAVADAGLSVAAVALGAVKQSLGEESPKTVDPITDLFPLRGAIVGANRVAELTENDRAMGRALVRGGA ADKLMRPGGIVDLLTAPGGLLDRVSAADSSSLERILAPGGLADRLLAEDGPLERMFAEDGIIERMFAEDG VIDKLLVKNGPLEQFTEAAEILSQLAPAVDKLTPTTDTLESAVVTLNKMVNSLSGITDRIPRRRSTRPAV RAKRTTDQAGVDE
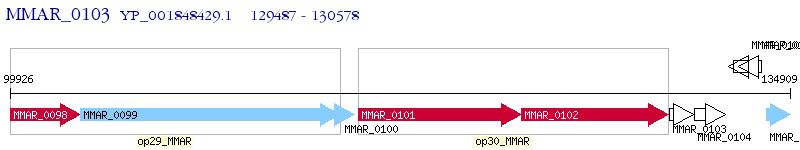
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0103 | - | - | 100% (363) | hypothetical protein MMAR_0103 |
| M. marinum M | MMAR_4943 | - | 1e-58 | 48.13% (241) | hypothetical protein MMAR_4943 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0779c | - | 9e-55 | 44.58% (240) | hypothetical protein Mb0779c |
| M. gilvum PYR-GCK | Mflv_1580 | - | 5e-55 | 46.12% (232) | hypothetical protein Mflv_1580 |
| M. tuberculosis H37Rv | Rv0756c | - | 9e-55 | 44.58% (240) | hypothetical protein Rv0756c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0672c | - | 1e-41 | 39.15% (235) | hypothetical protein MAB_0672c |
| M. avium 104 | MAV_0700 | - | 3e-61 | 50.42% (236) | hypothetical protein MAV_0700 |
| M. smegmatis MC2 155 | MSMEG_5873 | - | 4e-51 | 41.26% (269) | hypothetical protein MSMEG_5873 |
| M. thermoresistible (build 8) | TH_1249 | - | 7e-55 | 48.26% (230) | HYPOTHETICAL PROTEIN Rv0756c |
| M. ulcerans Agy99 | MUL_0462 | - | 8e-59 | 48.13% (241) | hypothetical protein MUL_0462 |
| M. vanbaalenii PYR-1 | Mvan_5176 | - | 9e-53 | 45.49% (233) | hypothetical protein Mvan_5176 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1580|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5176|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1249|M.thermoresistible__bu --------------------------------------------------
MSMEG_5873|M.smegmatis_MC2_155 --------------------------------------------------
Mb0779c|M.bovis_AF2122/97 --------------------------------------------------
Rv0756c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0462|M.ulcerans_Agy99 --------------------------------------------------
MAV_0700|M.avium_104 --------------------------------------------------
MMAR_0103|M.marinum_M MQQLGPKCARGQLSQPDRAWLDAARSSRSQPSTDRLSALKLSGSQLSATD
MAB_0672c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1580|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5176|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1249|M.thermoresistible__bu --------------------------------------------------
MSMEG_5873|M.smegmatis_MC2_155 --------------------------------------------------
Mb0779c|M.bovis_AF2122/97 --------------------------------------------------
Rv0756c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0462|M.ulcerans_Agy99 --------------------------------------------------
MAV_0700|M.avium_104 --------------------------------------------------
MMAR_0103|M.marinum_M ARAIGQSTQIFPRHVSKISLWSTSSGCNPGLKYTFALGAMPSKKWDTGQH
MAB_0672c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1580|M.gilvum_PYR-GCK ------------------------MNLAKSLTELAFAPMRAGLAVADAGI
Mvan_5176|M.vanbaalenii_PYR-1 ------------------------MNLGRSLTDLAFAPVRAGLAIAEVGL
TH_1249|M.thermoresistible__bu ------------------------MNLGKSLMQVATAPVRIGLAVADAGL
MSMEG_5873|M.smegmatis_MC2_155 ------------------------MNLGKTLVQVATAPVRIGIAIADAGL
Mb0779c|M.bovis_AF2122/97 ------------------------MNLGQTLVGIATWPARAGLAAADTGL
Rv0756c|M.tuberculosis_H37Rv ------------------------MNLGQTLVGIATWPARAGLAAADTGL
MUL_0462|M.ulcerans_Agy99 ------------------------MNVGQSLVAFATAPARAGLAAAEASL
MAV_0700|M.avium_104 ------------------------MNLAKNIVSAATAPARAGLAAADAGL
MMAR_0103|M.marinum_M DRAAQRFDGMEPGPRSAAEPYTCRVNISKAIVGAATAPIRAGVAVADAGL
MAB_0672c|M.abscessus_ATCC_199 ----------------------------MKLVSLITAPLKISLAVADTGL
: * : .:* *:..:
Mflv_1580|M.gilvum_PYR-GCK GVARSGIDIASRAMGDSAN--------------ATQTPTSFASMLGVQDA
Mvan_5176|M.vanbaalenii_PYR-1 GMASGAMDIAQRSLGDSGN-------------GARQAPTSFASMLGVQDA
TH_1249|M.thermoresistible__bu EVAAAGLGMAHRTLG-----------------EPTAGPSSVAGILGLEDT
MSMEG_5873|M.smegmatis_MC2_155 GIAGETLTAVQRTVTDS---------------NPLTSRSSLAQMLGLDDA
Mb0779c|M.bovis_AF2122/97 NMAGAAVDMAKQALGDA---------------GGASGSTSMANMLGIDDT
Rv0756c|M.tuberculosis_H37Rv NMAGAAVDMAKQALGDA---------------GGASGSTSMANMLGIDDT
MUL_0462|M.ulcerans_Agy99 NLAGAAVGLAKQALGET---------------STDAGNNAMASVLGIEDA
MAV_0700|M.avium_104 SVASAAVGVAKRALGDS---------------G-TAGTNAMTSMLGIDDA
MMAR_0103|M.marinum_M SVAAVALGAVKQSLGEE----------------SPKTVDPITDLFPLRGA
MAB_0672c|M.abscessus_ATCC_199 AVASRAIDTGREILAALPDGRPADDQGDAPGAGLVSAVESLFGMAGRKSP
:* : . : .. : ..
Mflv_1580|M.gilvum_PYR-GCK VERANRLAKLMDEDQPLGRALASGGPIDRLLQPGGVVDRLTAPG------
Mvan_5176|M.vanbaalenii_PYR-1 VDRANRLARLMDEDQPLGRALAPDGPIDRLLRPGGVVDRLTAPG------
TH_1249|M.thermoresistible__bu LERANRLAKLLDEDAPLGRALAPDGPLDRLMRPGGVIDRLTAPG------
MSMEG_5873|M.smegmatis_MC2_155 VERANRLARLMDDDAPLGRALAPDGPVNRLLRPGGLVDQLTAEGGLLDRL
Mb0779c|M.bovis_AF2122/97 IARANRLARLLDDDMPLGRAIAPNGPMDRMLRPGGVVDLLTQPG------
Rv0756c|M.tuberculosis_H37Rv IARANRLARLLDDDMPLGRAIAPNGPMDRMLRPGGVVDLLTQPG------
MUL_0462|M.ulcerans_Agy99 IVRANRLAKLLDEDAPLGRAIAPNGPMDRMLRPGGVVDLLTAPG------
MAV_0700|M.avium_104 IVRANRLARLLDDDAPLGRAVAPDGPLDRLLRPGGVVDMLTSEG------
MMAR_0103|M.marinum_M IVGANRVAELTENDRAMGRALVRGGAADKLMRPGGIVDLLTAPG------
MAB_0672c|M.abscessus_ATCC_199 LHSAMKVAELMEEDKPLGRALRAGGPVERIMAPGGLADRLTEPG------
: * ::*.* ::* .:***: .*. :::: ***: * ** *
Mflv_1580|M.gilvum_PYR-GCK ------------------------GALDRMTQDDGG-LMRAIEPGGLVDQ
Mvan_5176|M.vanbaalenii_PYR-1 ------------------------GVLDRLTEEEGG-LMRTLEPGGLVDQ
TH_1249|M.thermoresistible__bu ------------------------GVLDRLTDEEGG-LMRALEPGGLVDQ
MSMEG_5873|M.smegmatis_MC2_155 TAENGPVTRAVAPGGLVDQITMEGGLLDRLTTDDGA-LSRVIAPGGLADQ
Mb0779c|M.bovis_AF2122/97 ------------------------GLLDRLTAEGGA-MQRALQPGGLADQ
Rv0756c|M.tuberculosis_H37Rv ------------------------GLLDRLTAEGGA-MQRALQPGGLADQ
MUL_0462|M.ulcerans_Agy99 ------------------------GLLDRVTAEGGA-MHRALQPGGLADQ
MAV_0700|M.avium_104 ------------------------GLLDRLTAEGGA-LQRTLQPGGLADQ
MMAR_0103|M.marinum_M ------------------------GLLDRVSAADSSSLERILAPGGLADR
MAB_0672c|M.abscessus_ATCC_199 ------------------------GVLDRLFAKDGP-VDRLTAPDGAVDR
* ***: . : * *.* .*:
Mflv_1580|M.gilvum_PYR-GCK LLAEDGLVDRLLAEDGVADRLFAEGGVVDTLTARNGPLEQLAAVADTLNR
Mvan_5176|M.vanbaalenii_PYR-1 LLAEDGLVDRLLAEDGVADRLFAEGGVVDTLTARNGPLEQLAAVADTLNR
TH_1249|M.thermoresistible__bu LLAEDGLVERVLAEDGLADRLLAEGGLIDRLTARDGPLEQLADVADTLNR
MSMEG_5873|M.smegmatis_MC2_155 LLAHDGLIERVLREDGVADRLLAEGGLLDTLTDPDGPLLKLADVADTLNR
Mb0779c|M.bovis_AF2122/97 LLAEDGLIERVLSEDGLADRLLAEGGLIDKITAKDGPLEQLADVADTLAR
Rv0756c|M.tuberculosis_H37Rv LLAEDGLIERVLSEDGLADRLLAEGGLIDKITAKDGPLEQLADVADTLAR
MUL_0462|M.ulcerans_Agy99 LVAEDGLIERVLSEDGLADRLLSEGGLVDKLTAKNGPLEQLADVADTLAR
MAV_0700|M.avium_104 LVAEDGLIERLLAEDGLADRLLAEGGLIDKLTAKHGPLDQLADVADTLAR
MMAR_0103|M.marinum_M LLAEDGPLERMFAEDGIIERMFAEDGVIDKLLVKNGPLEQFTEAAEILSQ
MAB_0672c|M.abscessus_ATCC_199 ALAPGGLVDQLLAEDGILERLMREEGVLDKFTATDGPLQQLADLSEMLTK
:* .* ::::: ***: :*:: * *::* : .*** ::: :: * :
Mflv_1580|M.gilvum_PYR-GCK LAPGLEALEPTIEALRDAVIVLSQVVNPLSNIADRIPLPGRRIRNRPTTR
Mvan_5176|M.vanbaalenii_PYR-1 LAPGLEALEPTIEALREAVIVLSQVVNPLSNIADRIPLPGRRMRPRPTSR
TH_1249|M.thermoresistible__bu LAPGLEALEPTIAALREAVITLSQVVNPLSNIADRIPFP-RRPPRRMPPR
MSMEG_5873|M.smegmatis_MC2_155 LAPGLEALAPTIDALHDAVITLSQVVNPLSNIADRIPLPRRYGRRSTTPT
Mb0779c|M.bovis_AF2122/97 LTPGMEALEPAIATLQDAVIALTMVVNPLSSIAERIPLPGRRPARRSSSR
Rv0756c|M.tuberculosis_H37Rv LTPGMEALEPAIATLQDAVIALTMVVNPLSSIAERIPLPGRRPARRSSSR
MUL_0462|M.ulcerans_Agy99 LAPGMEALEPAIETLQDAVVALTMVVNPLSSIAERIPLPGRRPARRSSSR
MAV_0700|M.avium_104 LTPGMEALEPAIATLQDAVVALTMVVNPLSNIADRIPLPGRR---RPSSR
MMAR_0103|M.marinum_M LAPAVDKLTPTTDTLESAVVTLNKMVNSLSGITDRIPRRRST---RPAVR
MAB_0672c|M.abscessus_ATCC_199 AAPSIDALTPTVELLTDTVSALSSVMSPLGGFLPR----RRPARPSGSPR
:*.:: * *: * .:* .*. ::..*..: * .
Mflv_1580|M.gilvum_PYR-GCK PVPTQRIIESEE-
Mvan_5176|M.vanbaalenii_PYR-1 PVTSQRIVEADD-
TH_1249|M.thermoresistible__bu AVPSQRIIEPDDE
MSMEG_5873|M.smegmatis_MC2_155 PVVSQRIVDADE-
Mb0779c|M.bovis_AF2122/97 SVRSQRVVDSE--
Rv0756c|M.tuberculosis_H37Rv SVRSQRVVDSE--
MUL_0462|M.ulcerans_Agy99 PVRSQRVVDADE-
MAV_0700|M.avium_104 AVRSARVIDTDE-
MMAR_0103|M.marinum_M AKRTTDQAGVDE-
MAB_0672c|M.abscessus_ATCC_199 PARSERVIEGER-
. : :