For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNLGKTLVQVATAPVRIGIAIADAGLGIAGETLTAVQRTVTDSNPLTSRSSLAQMLGLDDAVERANRLAR LMDDDAPLGRALAPDGPVNRLLRPGGLVDQLTAEGGLLDRLTAENGPVTRAVAPGGLVDQITMEGGLLDR LTTDDGALSRVIAPGGLADQLLAHDGLIERVLREDGVADRLLAEGGLLDTLTDPDGPLLKLADVADTLNR LAPGLEALAPTIDALHDAVITLSQVVNPLSNIADRIPLPRRYGRRSTTPTPVVSQRIVDADE
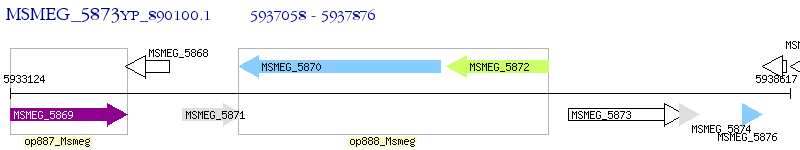
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5873 | - | - | 100% (272) | hypothetical protein MSMEG_5873 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0779c | - | 4e-75 | 54.24% (271) | hypothetical protein Mb0779c |
| M. gilvum PYR-GCK | Mflv_1580 | - | 2e-77 | 56.41% (273) | hypothetical protein Mflv_1580 |
| M. tuberculosis H37Rv | Rv0756c | - | 4e-75 | 54.24% (271) | hypothetical protein Rv0756c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0672c | - | 1e-42 | 36.07% (280) | hypothetical protein MAB_0672c |
| M. marinum M | MMAR_4943 | - | 1e-73 | 52.21% (272) | hypothetical protein MMAR_4943 |
| M. avium 104 | MAV_0700 | - | 5e-75 | 56.25% (272) | hypothetical protein MAV_0700 |
| M. thermoresistible (build 8) | TH_1249 | - | 8e-82 | 61.03% (272) | HYPOTHETICAL PROTEIN Rv0756c |
| M. ulcerans Agy99 | MUL_0462 | - | 1e-73 | 52.21% (272) | hypothetical protein MUL_0462 |
| M. vanbaalenii PYR-1 | Mvan_5176 | - | 2e-78 | 57.30% (274) | hypothetical protein Mvan_5176 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0779c|M.bovis_AF2122/97 MNLGQTLVGIATWPARAGLAAADTGLNMAGAAVDMAKQALGDAGG-----
Rv0756c|M.tuberculosis_H37Rv MNLGQTLVGIATWPARAGLAAADTGLNMAGAAVDMAKQALGDAGG-----
MMAR_4943|M.marinum_M MNVGQSLVAFATAPARAGLAAAEASLNLAGAAVGLAKQALGETST-----
MUL_0462|M.ulcerans_Agy99 MNVGQSLVAFATAPARAGLAAAEASLNLAGAAVGLAKQALGETST-----
MAV_0700|M.avium_104 MNLAKNIVSAATAPARAGLAAADAGLSVASAAVGVAKRALGDSG------
Mflv_1580|M.gilvum_PYR-GCK MNLAKSLTELAFAPMRAGLAVADAGIGVARSGIDIASRAMGDSAN-----
Mvan_5176|M.vanbaalenii_PYR-1 MNLGRSLTDLAFAPVRAGLAIAEVGLGMASGAMDIAQRSLGDSGNG----
TH_1249|M.thermoresistible__bu MNLGKSLMQVATAPVRIGLAVADAGLEVAAAGLGMAHRTLG----E----
MSMEG_5873|M.smegmatis_MC2_155 MNLGKTLVQVATAPVRIGIAIADAGLGIAGETLTAVQRTVTDSNP-----
MAB_0672c|M.abscessus_ATCC_199 ----MKLVSLITAPLKISLAVADTGLAVASRAIDTGREILAALPDGRPAD
.: * : .:* *:..: :* : . :
Mb0779c|M.bovis_AF2122/97 ----------ASGSTSMANMLGIDDTIARANRLARLLDDDMPLGRAIAPN
Rv0756c|M.tuberculosis_H37Rv ----------ASGSTSMANMLGIDDTIARANRLARLLDDDMPLGRAIAPN
MMAR_4943|M.marinum_M ----------DAGNNAMASVLGIEDAIVRANRLAKLLDEDAPLGRAIAPN
MUL_0462|M.ulcerans_Agy99 ----------DAGNNAMASVLGIEDAIVRANRLAKLLDEDAPLGRAIAPN
MAV_0700|M.avium_104 ----------TAGTNAMTSMLGIDDAIVRANRLARLLDDDAPLGRAVAPD
Mflv_1580|M.gilvum_PYR-GCK ---------ATQTPTSFASMLGVQDAVERANRLAKLMDEDQPLGRALASG
Mvan_5176|M.vanbaalenii_PYR-1 ---------ARQAPTSFASMLGVQDAVDRANRLARLMDEDQPLGRALAPD
TH_1249|M.thermoresistible__bu ---------PTAGPSSVAGILGLEDTLERANRLAKLLDEDAPLGRALAPD
MSMEG_5873|M.smegmatis_MC2_155 ----------LTSRSSLAQMLGLDDAVERANRLARLMDDDAPLGRALAPD
MAB_0672c|M.abscessus_ATCC_199 DQGDAPGAGLVSAVESLFGMAGRKSPLHSAMKVAELMEEDKPLGRALRAG
:. : * ...: * ::*.*:::* *****: ..
Mb0779c|M.bovis_AF2122/97 GPMDRMLRPGGVVDLLTQPGGLLDRLTAE---------------------
Rv0756c|M.tuberculosis_H37Rv GPMDRMLRPGGVVDLLTQPGGLLDRLTAE---------------------
MMAR_4943|M.marinum_M GPMDRMLRPGGVVDLLTAPGGLLDRVTAE---------------------
MUL_0462|M.ulcerans_Agy99 GPMDRMLRPGGVVDLLTAPGGLLDRVTAE---------------------
MAV_0700|M.avium_104 GPLDRLLRPGGVVDMLTSEGGLLDRLTAE---------------------
Mflv_1580|M.gilvum_PYR-GCK GPIDRLLQPGGVVDRLTAPGGALDRMTQD---------------------
Mvan_5176|M.vanbaalenii_PYR-1 GPIDRLLRPGGVVDRLTAPGGVLDRLTEE---------------------
TH_1249|M.thermoresistible__bu GPLDRLMRPGGVIDRLTAPGGVLDRLTDE---------------------
MSMEG_5873|M.smegmatis_MC2_155 GPVNRLLRPGGLVDQLTAEGGLLDRLTAENGPVTRAVAPGGLVDQITMEG
MAB_0672c|M.abscessus_ATCC_199 GPVERIMAPGGLADRLTEPGGVLDRLFAK---------------------
**::*:: ***: * ** ** ***: .
Mb0779c|M.bovis_AF2122/97 ---------GGAMQRALQPGGLADQLLAEDGLIERVLSEDGLADRLLAEG
Rv0756c|M.tuberculosis_H37Rv ---------GGAMQRALQPGGLADQLLAEDGLIERVLSEDGLADRLLAEG
MMAR_4943|M.marinum_M ---------GGAMHRALQPGGLADQLVAEDGLIERVLSEDGLADRLLSEG
MUL_0462|M.ulcerans_Agy99 ---------GGAMHRALQPGGLADQLVAEDGLIERVLSEDGLADRLLSEG
MAV_0700|M.avium_104 ---------GGALQRTLQPGGLADQLVAEDGLIERLLAEDGLADRLLAEG
Mflv_1580|M.gilvum_PYR-GCK ---------DGGLMRAIEPGGLVDQLLAEDGLVDRLLAEDGVADRLFAEG
Mvan_5176|M.vanbaalenii_PYR-1 ---------EGGLMRTLEPGGLVDQLLAEDGLVDRLLAEDGVADRLFAEG
TH_1249|M.thermoresistible__bu ---------EGGLMRALEPGGLVDQLLAEDGLVERVLAEDGLADRLLAEG
MSMEG_5873|M.smegmatis_MC2_155 GLLDRLTTDDGALSRVIAPGGLADQLLAHDGLIERVLREDGVADRLLAEG
MAB_0672c|M.abscessus_ATCC_199 ---------DGPVDRLTAPDGAVDRALAPGGLVDQLLAEDGILERLMREE
* : * *.* .*: :* .**::::* ***: :**: *
Mb0779c|M.bovis_AF2122/97 GLIDKITAKDGPLEQLADVADTLARLTPGMEALEPAIATLQDAVIALTMV
Rv0756c|M.tuberculosis_H37Rv GLIDKITAKDGPLEQLADVADTLARLTPGMEALEPAIATLQDAVIALTMV
MMAR_4943|M.marinum_M GLVDKLTAKNGPLEQLADVADTLARLAPGMEALEPAIETLQDAVVALTMV
MUL_0462|M.ulcerans_Agy99 GLVDKLTAKNGPLEQLADVADTLARLAPGMEALEPAIETLQDAVVALTMV
MAV_0700|M.avium_104 GLIDKLTAKHGPLDQLADVADTLARLTPGMEALEPAIATLQDAVVALTMV
Mflv_1580|M.gilvum_PYR-GCK GVVDTLTARNGPLEQLAAVADTLNRLAPGLEALEPTIEALRDAVIVLSQV
Mvan_5176|M.vanbaalenii_PYR-1 GVVDTLTARNGPLEQLAAVADTLNRLAPGLEALEPTIEALREAVIVLSQV
TH_1249|M.thermoresistible__bu GLIDRLTARDGPLEQLADVADTLNRLAPGLEALEPTIAALREAVITLSQV
MSMEG_5873|M.smegmatis_MC2_155 GLLDTLTDPDGPLLKLADVADTLNRLAPGLEALAPTIDALHDAVITLSQV
MAB_0672c|M.abscessus_ATCC_199 GVLDKFTATDGPLQQLADLSEMLTKAAPSIDALTPTVELLTDTVSALSSV
*::* :* .*** :** ::: * : :*.::** *:: * ::* .*: *
Mb0779c|M.bovis_AF2122/97 VNPLSSIAERIPLPGRRPARRSSSRSVRSQRVVDSE--
Rv0756c|M.tuberculosis_H37Rv VNPLSSIAERIPLPGRRPARRSSSRSVRSQRVVDSE--
MMAR_4943|M.marinum_M VNPLSSIAERIPLPGRRPARRSSSRPVRSQRVVDADE-
MUL_0462|M.ulcerans_Agy99 VNPLSSIAERIPLPGRRPARRSSSRPVRSQRVVDADE-
MAV_0700|M.avium_104 VNPLSNIADRIPLPGRR---RPSSRAVRSARVIDTDE-
Mflv_1580|M.gilvum_PYR-GCK VNPLSNIADRIPLPGRRIRNRPTTRPVPTQRIIESEE-
Mvan_5176|M.vanbaalenii_PYR-1 VNPLSNIADRIPLPGRRMRPRPTSRPVTSQRIVEADD-
TH_1249|M.thermoresistible__bu VNPLSNIADRIPFP-RRPPRRMPPRAVPSQRIIEPDDE
MSMEG_5873|M.smegmatis_MC2_155 VNPLSNIADRIPLPRRYGRRSTTPTPVVSQRIVDADE-
MAB_0672c|M.abscessus_ATCC_199 MSPLGGFLPR----RRPARPSGSPRPARSERVIEGER-
:.**..: * * .. .. : *::: :