For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALAFSAGTVDDLLREAYTAIREQGQPVLPTRAPTRELIGVSLELTEPRCRLSRTEARRLSRTGIAQLAWY LSGTTNPEMILHWVPNHRNELEADGTIHGAYGPRMFGTGDNAQFCTVLELLKTSSSSRRAVIQLFDATDL TGPGRYKDVPCTNTLQFFVRDSRLHLMVTMRSNDAYIGLPLDVFAFTMLQELMAGELGIGIGRYIHSVGS LHIYEYNKEAIIDFLAEGWQSTDTPMPAMPAQSLTRHVPQFLASELQIRSGTPFADLTLPSDPYWADLAR MLALDSARGRKAHDEVAAVKESIAAPFFSEFLRRD*
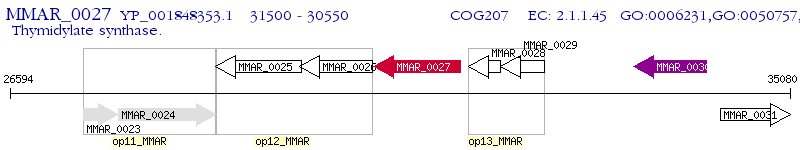
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0027 | thyA_1 | - | 100% (316) | thymidylate synthase, ThyA_1 |
| M. marinum M | MMAR_1956 | thyA | 9e-14 | 26.16% (172) | thymidylate synthase ThyA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2786c | thyA | 1e-14 | 27.71% (166) | thymidylate synthase |
| M. gilvum PYR-GCK | Mflv_4007 | thyA | 2e-14 | 22.94% (231) | thymidylate synthase |
| M. tuberculosis H37Rv | Rv2764c | thyA | 1e-14 | 27.71% (166) | thymidylate synthase |
| M. leprae Br4923 | MLBr_01519 | thyA | 5e-14 | 25.60% (168) | thymidylate synthase |
| M. abscessus ATCC 19977 | MAB_3091c | - | 7e-15 | 26.22% (225) | thymidylate synthase (ThyA) |
| M. avium 104 | MAV_3652 | thyA | 1e-14 | 26.79% (168) | thymidylate synthase |
| M. smegmatis MC2 155 | MSMEG_2670 | thyA | 7e-15 | 25.56% (223) | thymidylate synthase |
| M. thermoresistible (build 8) | TH_0692 | thyA | 5e-16 | 26.46% (223) | PROBABLE THYMIDYLATE SYNTHASE THYA (TS) (TSASE) |
| M. ulcerans Agy99 | MUL_2178 | thyA | 2e-14 | 26.16% (172) | thymidylate synthase |
| M. vanbaalenii PYR-1 | Mvan_2380 | thyA | 4e-14 | 26.01% (173) | thymidylate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2786c|M.bovis_AF2122/97 -------MTPYEDLLRFVLETGTPKSD---RTGTGTRSLFGQQMRYDLSA
Rv2764c|M.tuberculosis_H37Rv -------MTPYEDLLRFVLETGTPKSD---RTGTGTRSLFGQQMRYDLSA
MUL_2178|M.ulcerans_Agy99 ----MPIATPYEDLLRRVLETGTAKSD---RTGTGTRSLFGQQIRYDLGC
MLBr_01519|M.leprae_Br4923 ----MLIATPYEDLLRLVLDCGVAKMD---RTGTGTRSLFGQQLRYDLSV
MAV_3652|M.avium_104 ----MPIPTPYEDLLRLVLDRGTAKSD---RTGTGTRSLFGQQLRYDLSA
Mflv_4007|M.gilvum_PYR-GCK ----MPTATPYEDLLRLVLDTGTPKSD---RTGTGTRSLFGHQMRYDLNA
Mvan_2380|M.vanbaalenii_PYR-1 ----MPIATPYEDLLRLVLDTGTTKSD---RTGTGTRSLFGHQMRYDLNA
MSMEG_2670|M.smegmatis_MC2_155 ----MPIDTPYEDLLRLVTERGTPKSD---RTGTGTRSLFGHQMRYDLSA
TH_0692|M.thermoresistible__bu VPINGSIPTPYEDLLRLVLERGTPKAD---RTGTGTRSLFGHQMRYDLSA
MAB_3091c|M.abscessus_ATCC_199 ----MSLPTPYEDLLRLVLEEGTPKAD---RTGTGTRSVFGHQLRYRLED
MMAR_0027|M.marinum_M -MALAFSAGTVDDLLREAYTAIREQGQPVLPTRAPTRELIGVSLELTEPR
. :**** . : : * : **.::* .:.
Mb2786c|M.bovis_AF2122/97 GFPLLTTKKVHFKSVAYELLWFLRGDSNIGWLHEHGVTIWDEWASDTGEL
Rv2764c|M.tuberculosis_H37Rv GFPLLTTKKVHFKSVAYELLWFLRGDSNIGWLHEHGVTIWDEWASDTGEL
MUL_2178|M.ulcerans_Agy99 GFPLLTTKKVHFKSVVYELLWFLRGDSNVGWLQQHGVTIWDEWASETGDL
MLBr_01519|M.leprae_Br4923 GFPLVTTKKVHFKSVVYELLWFLRGDSNVAWLHEHGVNIWDEWASSTGDL
MAV_3652|M.avium_104 GFPLITTKKVHLKSVVYELLWFLRGDSNVDWLHRHGVTIWDEWASDTGDL
Mflv_4007|M.gilvum_PYR-GCK GFPLITTKKVHTKSIVYELLWFLRGDSNVRWLQERGVTIWDEWASPTGDL
Mvan_2380|M.vanbaalenii_PYR-1 GFPLITTKKVHTKSIVYELLWFLRGDSNVRWLQEHGVTIWDEWASPTGDL
MSMEG_2670|M.smegmatis_MC2_155 GFPLITTKKVHTKSVIYELLWFLRGDSNVRWLQEHGVTIWDEWASETGDL
TH_0692|M.thermoresistible__bu GFPLITTKKVHTKSVIYELLWFLRGDSNVRWLQDRGVTIWDEWADDDGEL
MAB_3091c|M.abscessus_ATCC_199 GFPLITTKKVHLKSVIYELLWFLRGESNVRWLQERGVTIWDEWASEAGEL
MMAR_0027|M.marinum_M CRLSRTEARRLSRTGIAQLAWYLSGTTNPEMILHWVPNHRNELEAD-GTI
* : :: :* *:* * :* : . :* * :
Mb2786c|M.bovis_AF2122/97 GPIYGVQWRSWPAPSGEHIDQISAALDLLRTDPDSRRIIVSAWNVGEIE-
Rv2764c|M.tuberculosis_H37Rv GPIYGVQWRSWPAPSGEHIDQISAALDLLRTDPDSRRIIVSAWNVGEIE-
MUL_2178|M.ulcerans_Agy99 GPIYGVQWRSWPTPSGEHIDQISAALELLRTDPDSRRIIVSAWNVGQIP-
MLBr_01519|M.leprae_Br4923 GPIYGVQWRSWPTSSGDYIDQISTALDLLRTEPDSRRIIVSAWNVGEIP-
MAV_3652|M.avium_104 GPIYGVQWRSWPTPSGEHIDQISAALDLLRTDPDSRRIIVSAWNVGEIP-
Mflv_4007|M.gilvum_PYR-GCK GPVYGVQWRSWPTPSGEHIDQISAALDLLKRDPDSRRNIVSAWNVGEIP-
Mvan_2380|M.vanbaalenii_PYR-1 GPVYGVQWRSWPTPSGAHIDQISAALDLLRRDPDSRRNIVSAWNVGEIP-
MSMEG_2670|M.smegmatis_MC2_155 GPIYGVQWRSWPTPSGEHIDQISSALELLKSDPDSRRNIVSAWNVGEIP-
TH_0692|M.thermoresistible__bu GPVYGVQWRSWPTPSGEHVDQISAALELLRTDPDSRRIIVSAWNVADLD-
MAB_3091c|M.abscessus_ATCC_199 GPVYGVQWRAWPTPSGEHIDQISAALDLLRTNPDSRRNIVSAWNVGEIP-
MMAR_0027|M.marinum_M HGAYGP--RMFGTGDN---AQFCTVLELLKTSSSSRRAVIQLFDATDLTG
** * : : .. *:.:.*:**: ...*** ::. ::. ::
Mb2786c|M.bovis_AF2122/97 --RMALPPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHM
Rv2764c|M.tuberculosis_H37Rv --RMALPPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHM
MUL_2178|M.ulcerans_Agy99 --QMALPPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHM
MLBr_01519|M.leprae_Br4923 --QMALPPCHAFFQFYVAQGRLSCQLYQRSADMFLGVPFNIASYALLTHM
MAV_3652|M.avium_104 --RMALPPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHM
Mflv_4007|M.gilvum_PYR-GCK --QMALPPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHM
Mvan_2380|M.vanbaalenii_PYR-1 --QMALPPCHAFFQFYVSGGRLSCQLYQRSADLFLGVPFNIASYALLTHM
MSMEG_2670|M.smegmatis_MC2_155 --QMALPPCHAFFQFYVADGKLSCQLYQRSADLFLGVPFNIASYALLTHM
TH_0692|M.thermoresistible__bu --EMALPPCHTLFQFYVAGGRLSCQLYQRSADLFLGVPFNIASYALLTHM
MAB_3091c|M.abscessus_ATCC_199 --QMALPPCHAFFQFYVADGKLSCQLYQRSADLFLGVPFNIASYALLTHM
MMAR_0027|M.marinum_M PGRYKDVPCTNTLQFFVRDSRLHLMVTMRSNDAYIGLPLDVFAFTMLQEL
. ** :**:* .:* : ** * ::*:*::: ::::* .:
Mb2786c|M.bovis_AF2122/97 MAAQAGLSVGEFIWTGGDCHIYDNHVEQVRLQLSR---------------
Rv2764c|M.tuberculosis_H37Rv MAAQAGLSVGEFIWTGGDCHIYDNHVEQVRLQLSR---------------
MUL_2178|M.ulcerans_Agy99 MAAQAGLSVGEFIWTGGDCHIYDNHVEQVTEQLRR---------------
MLBr_01519|M.leprae_Br4923 MAAQSGLLVGEFVWTGGDCHIYDNHVEQVQLQLSR---------------
MAV_3652|M.avium_104 MAAQAGLSVGEFVWTGGDCHIYDNHVEQVRLQLSR---------------
Mflv_4007|M.gilvum_PYR-GCK MAAQAGLGVGEFIWTGGDCHIYDNHVEQVTEQLSR---------------
Mvan_2380|M.vanbaalenii_PYR-1 MAAQAGLGVGEFIWTGGDCHIYDNHVEQVTEQLSR---------------
MSMEG_2670|M.smegmatis_MC2_155 MAAQAGLDVGEFIWTGGDCHIYDNHTEQVALQLSR---------------
TH_0692|M.thermoresistible__bu MAAQAGLQVGEFIWTGGDCHIYDNHVEQVRLQLSR---------------
MAB_3091c|M.abscessus_ATCC_199 MAAQAGLGVGDFVWTGGDCHIYDNHVEQVTEQLSR---------------
MMAR_0027|M.marinum_M MAGELGIGIGRYIHSVGSLHIYEYNKEAIIDFLAEGWQSTDTPMPAMPAQ
**.: *: :* :: : *. ***: : * : * .
Mb2786c|M.bovis_AF2122/97 -EPRPYPKLLLADRDSIFEYTYEDIVVKN---------------------
Rv2764c|M.tuberculosis_H37Rv -EPRPYPKLLLADRDSIFEYTYEDIVVKN---------------------
MUL_2178|M.ulcerans_Agy99 -EPRPYPKLSLSQRDSIFDYTYEDVVVQD---------------------
MLBr_01519|M.leprae_Br4923 -EPRAYPELFLAQRDSIFDYVYEDVVVTN---------------------
MAV_3652|M.avium_104 -EPRPYPELVLAHRDSIFDYTYDDVVVHN---------------------
Mflv_4007|M.gilvum_PYR-GCK -DPRPYPELVLAPRDSIFDYTYEDVVIKN---------------------
Mvan_2380|M.vanbaalenii_PYR-1 -EPRPYPELVLAQRDSLFDYIYEDVVIKN---------------------
MSMEG_2670|M.smegmatis_MC2_155 -EPRPYPELVLAPRDSIFDYTYEDIAIVN---------------------
TH_0692|M.thermoresistible__bu -DPRPYPELELAPRDSIFDYTFEDVVIRN---------------------
MAB_3091c|M.abscessus_ATCC_199 -DPYPYPTLVLAQRDSIFDYRYEDITVEN---------------------
MMAR_0027|M.marinum_M SLTRHVPQFLASELQIRSGTPFADLTLPSDPYWADLARMLALDSARGRKA
. * : : : : *:.: .
Mb2786c|M.bovis_AF2122/97 YDPHPAIKAPVAV----------
Rv2764c|M.tuberculosis_H37Rv YDPHPAIKAPVAV----------
MUL_2178|M.ulcerans_Agy99 YDPHPAIKAPVAV----------
MLBr_01519|M.leprae_Br4923 YDPHPAIKAPVAI----------
MAV_3652|M.avium_104 YDPHPAIKAPVAV----------
Mflv_4007|M.gilvum_PYR-GCK YEPHPAIKAPVAI----------
Mvan_2380|M.vanbaalenii_PYR-1 YDPHPAIKAPVAV----------
MSMEG_2670|M.smegmatis_MC2_155 YDPHPAIKAPVAV----------
TH_0692|M.thermoresistible__bu YDPHPAIKAPVAV----------
MAB_3091c|M.abscessus_ATCC_199 YQHHAAIKAPVAV----------
MMAR_0027|M.marinum_M HDEVAAVKESIAAPFFSEFLRRD
:: .*:* .:*