For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLPTPYEDLLRLVLEEGTPKADRTGTGTRSVFGHQLRYRLEDGFPLITTKKVHLKSVIYELLWFLRGESN VRWLQERGVTIWDEWASEAGELGPVYGVQWRAWPTPSGEHIDQISAALDLLRTNPDSRRNIVSAWNVGEI PQMALPPCHAFFQFYVADGKLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAGLGVGDFVWTGGDCHI YDNHVEQVTEQLSRDPYPYPTLVLAQRDSIFDYRYEDITVENYQHHAAIKAPVAV*
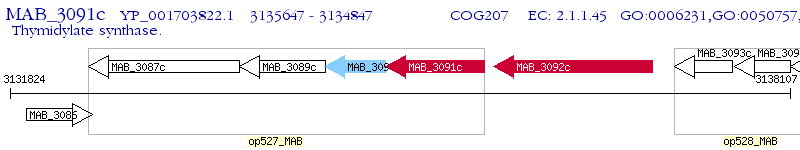
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3091c | - | - | 100% (266) | thymidylate synthase (ThyA) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2786c | thyA | 1e-133 | 83.21% (262) | thymidylate synthase |
| M. gilvum PYR-GCK | Mflv_4007 | thyA | 1e-141 | 86.09% (266) | thymidylate synthase |
| M. tuberculosis H37Rv | Rv2764c | thyA | 1e-133 | 83.21% (262) | thymidylate synthase |
| M. leprae Br4923 | MLBr_01519 | thyA | 1e-128 | 80.08% (266) | thymidylate synthase |
| M. marinum M | MMAR_1956 | thyA | 1e-133 | 82.33% (266) | thymidylate synthase ThyA |
| M. avium 104 | MAV_3652 | thyA | 1e-136 | 84.21% (266) | thymidylate synthase |
| M. smegmatis MC2 155 | MSMEG_2670 | thyA | 1e-137 | 84.96% (266) | thymidylate synthase |
| M. thermoresistible (build 8) | TH_0692 | thyA | 1e-134 | 83.02% (265) | PROBABLE THYMIDYLATE SYNTHASE THYA (TS) (TSASE) |
| M. ulcerans Agy99 | MUL_2178 | thyA | 1e-134 | 82.71% (266) | thymidylate synthase |
| M. vanbaalenii PYR-1 | Mvan_2380 | thyA | 1e-138 | 84.59% (266) | thymidylate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3091c|M.abscessus_ATCC_199 ----MSLPTPYEDLLRLVLEEGTPKADRTGTGTRSVFGHQLRYRLEDGFP
TH_0692|M.thermoresistible__bu VPINGSIPTPYEDLLRLVLERGTPKADRTGTGTRSLFGHQMRYDLSAGFP
Mflv_4007|M.gilvum_PYR-GCK ----MPTATPYEDLLRLVLDTGTPKSDRTGTGTRSLFGHQMRYDLNAGFP
Mvan_2380|M.vanbaalenii_PYR-1 ----MPIATPYEDLLRLVLDTGTTKSDRTGTGTRSLFGHQMRYDLNAGFP
MSMEG_2670|M.smegmatis_MC2_155 ----MPIDTPYEDLLRLVTERGTPKSDRTGTGTRSLFGHQMRYDLSAGFP
Mb2786c|M.bovis_AF2122/97 -------MTPYEDLLRFVLETGTPKSDRTGTGTRSLFGQQMRYDLSAGFP
Rv2764c|M.tuberculosis_H37Rv -------MTPYEDLLRFVLETGTPKSDRTGTGTRSLFGQQMRYDLSAGFP
MMAR_1956|M.marinum_M ----MPIATPYEDLLRRVLETGTAKSDRTGTGTRSLFGQQIRYDLGCGFP
MUL_2178|M.ulcerans_Agy99 ----MPIATPYEDLLRRVLETGTAKSDRTGTGTRSLFGQQIRYDLGCGFP
MLBr_01519|M.leprae_Br4923 ----MLIATPYEDLLRLVLDCGVAKMDRTGTGTRSLFGQQLRYDLSVGFP
MAV_3652|M.avium_104 ----MPIPTPYEDLLRLVLDRGTAKSDRTGTGTRSLFGQQLRYDLSAGFP
******** * : *..* *********:**:*:** * ***
MAB_3091c|M.abscessus_ATCC_199 LITTKKVHLKSVIYELLWFLRGESNVRWLQERGVTIWDEWASEAGELGPV
TH_0692|M.thermoresistible__bu LITTKKVHTKSVIYELLWFLRGDSNVRWLQDRGVTIWDEWADDDGELGPV
Mflv_4007|M.gilvum_PYR-GCK LITTKKVHTKSIVYELLWFLRGDSNVRWLQERGVTIWDEWASPTGDLGPV
Mvan_2380|M.vanbaalenii_PYR-1 LITTKKVHTKSIVYELLWFLRGDSNVRWLQEHGVTIWDEWASPTGDLGPV
MSMEG_2670|M.smegmatis_MC2_155 LITTKKVHTKSVIYELLWFLRGDSNVRWLQEHGVTIWDEWASETGDLGPI
Mb2786c|M.bovis_AF2122/97 LLTTKKVHFKSVAYELLWFLRGDSNIGWLHEHGVTIWDEWASDTGELGPI
Rv2764c|M.tuberculosis_H37Rv LLTTKKVHFKSVAYELLWFLRGDSNIGWLHEHGVTIWDEWASDTGELGPI
MMAR_1956|M.marinum_M LLTTKKVHFKSVVYELLWFLRGDSNVGWLQQHGVTIWDEWASETGDLGPI
MUL_2178|M.ulcerans_Agy99 LLTTKKVHFKSVVYELLWFLRGDSNVGWLQQHGVTIWDEWASETGDLGPI
MLBr_01519|M.leprae_Br4923 LVTTKKVHFKSVVYELLWFLRGDSNVAWLHEHGVNIWDEWASSTGDLGPI
MAV_3652|M.avium_104 LITTKKVHLKSVVYELLWFLRGDSNVDWLHRHGVTIWDEWASDTGDLGPI
*:****** **: *********:**: **: :**.******. *:***:
MAB_3091c|M.abscessus_ATCC_199 YGVQWRAWPTPSGEHIDQISAALDLLRTNPDSRRNIVSAWNVGEIPQMAL
TH_0692|M.thermoresistible__bu YGVQWRSWPTPSGEHVDQISAALELLRTDPDSRRIIVSAWNVADLDEMAL
Mflv_4007|M.gilvum_PYR-GCK YGVQWRSWPTPSGEHIDQISAALDLLKRDPDSRRNIVSAWNVGEIPQMAL
Mvan_2380|M.vanbaalenii_PYR-1 YGVQWRSWPTPSGAHIDQISAALDLLRRDPDSRRNIVSAWNVGEIPQMAL
MSMEG_2670|M.smegmatis_MC2_155 YGVQWRSWPTPSGEHIDQISSALELLKSDPDSRRNIVSAWNVGEIPQMAL
Mb2786c|M.bovis_AF2122/97 YGVQWRSWPAPSGEHIDQISAALDLLRTDPDSRRIIVSAWNVGEIERMAL
Rv2764c|M.tuberculosis_H37Rv YGVQWRSWPAPSGEHIDQISAALDLLRTDPDSRRIIVSAWNVGEIERMAL
MMAR_1956|M.marinum_M YGVQWRSWPTPSGEHIDQISAALELLRTDPDSRRIIVSAWNVGQIPQMAL
MUL_2178|M.ulcerans_Agy99 YGVQWRSWPTPSGEHIDQISAALELLRTDPDSRRIIVSAWNVGQIPQMAL
MLBr_01519|M.leprae_Br4923 YGVQWRSWPTSSGDYIDQISTALDLLRTEPDSRRIIVSAWNVGEIPQMAL
MAV_3652|M.avium_104 YGVQWRSWPTPSGEHIDQISAALDLLRTDPDSRRIIVSAWNVGEIPRMAL
******:**:.** ::****:**:**: :***** *******.:: .***
MAB_3091c|M.abscessus_ATCC_199 PPCHAFFQFYVADGKLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
TH_0692|M.thermoresistible__bu PPCHTLFQFYVAGGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
Mflv_4007|M.gilvum_PYR-GCK PPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
Mvan_2380|M.vanbaalenii_PYR-1 PPCHAFFQFYVSGGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
MSMEG_2670|M.smegmatis_MC2_155 PPCHAFFQFYVADGKLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
Mb2786c|M.bovis_AF2122/97 PPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
Rv2764c|M.tuberculosis_H37Rv PPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
MMAR_1956|M.marinum_M PPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
MUL_2178|M.ulcerans_Agy99 PPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
MLBr_01519|M.leprae_Br4923 PPCHAFFQFYVAQGRLSCQLYQRSADMFLGVPFNIASYALLTHMMAAQSG
MAV_3652|M.avium_104 PPCHAFFQFYVADGRLSCQLYQRSADLFLGVPFNIASYALLTHMMAAQAG
****::*****: *:***********:*********************:*
MAB_3091c|M.abscessus_ATCC_199 LGVGDFVWTGGDCHIYDNHVEQVTEQLSRDPYPYPTLVLAQRDSIFDYRY
TH_0692|M.thermoresistible__bu LQVGEFIWTGGDCHIYDNHVEQVRLQLSRDPRPYPELELAPRDSIFDYTF
Mflv_4007|M.gilvum_PYR-GCK LGVGEFIWTGGDCHIYDNHVEQVTEQLSRDPRPYPELVLAPRDSIFDYTY
Mvan_2380|M.vanbaalenii_PYR-1 LGVGEFIWTGGDCHIYDNHVEQVTEQLSREPRPYPELVLAQRDSLFDYIY
MSMEG_2670|M.smegmatis_MC2_155 LDVGEFIWTGGDCHIYDNHTEQVALQLSREPRPYPELVLAPRDSIFDYTY
Mb2786c|M.bovis_AF2122/97 LSVGEFIWTGGDCHIYDNHVEQVRLQLSREPRPYPKLLLADRDSIFEYTY
Rv2764c|M.tuberculosis_H37Rv LSVGEFIWTGGDCHIYDNHVEQVRLQLSREPRPYPKLLLADRDSIFEYTY
MMAR_1956|M.marinum_M LSVGEFIWTGGDCHIYGNHVEQVTEQLRREPRPYPKLSLSQRDSIFDYTY
MUL_2178|M.ulcerans_Agy99 LSVGEFIWTGGDCHIYDNHVEQVTEQLRREPRPYPKLSLSQRDSIFDYTY
MLBr_01519|M.leprae_Br4923 LLVGEFVWTGGDCHIYDNHVEQVQLQLSREPRAYPELFLAQRDSIFDYVY
MAV_3652|M.avium_104 LSVGEFVWTGGDCHIYDNHVEQVRLQLSREPRPYPELVLAHRDSIFDYTY
* **:*:*********.**.*** ** *:* .** * *: ***:*:* :
MAB_3091c|M.abscessus_ATCC_199 EDITVENYQHHAAIKAPVAV
TH_0692|M.thermoresistible__bu EDVVIRNYDPHPAIKAPVAV
Mflv_4007|M.gilvum_PYR-GCK EDVVIKNYEPHPAIKAPVAI
Mvan_2380|M.vanbaalenii_PYR-1 EDVVIKNYDPHPAIKAPVAV
MSMEG_2670|M.smegmatis_MC2_155 EDIAIVNYDPHPAIKAPVAV
Mb2786c|M.bovis_AF2122/97 EDIVVKNYDPHPAIKAPVAV
Rv2764c|M.tuberculosis_H37Rv EDIVVKNYDPHPAIKAPVAV
MMAR_1956|M.marinum_M EDVVVQDYDPHPAIKAPVAV
MUL_2178|M.ulcerans_Agy99 EDVVVQDYDPHPAIKAPVAV
MLBr_01519|M.leprae_Br4923 EDVVVTNYDPHPAIKAPVAI
MAV_3652|M.avium_104 DDVVVHNYDPHPAIKAPVAV
:*:.: :*: *.*******: