For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLGAERERRLTGWLRGRLPDADDVRVEGIDRVSFGHSAEMMTMSVIATRRGRDERRDVVLRLRPSPPALL EPYDLARQFGILRALADTDVRAPRALWLEPTGDVLGRPFFVMERVAGEVYEMQPPADASDDTVARMCESL AEQLAAIHAVDLTRTGLAPLDDGAGHLDRELGHWAAEMNRVKRGPLPALERLHRALLSGKPAPCPRITLV HGDAKPGNFAFTGGEVSAVFDWEMTTVGDPLTDIGWLEMLWMQPVGINSHPAALPIDALLAHYQSASGIE PVNRPWYRAFNAYKMAVICLIGAMLVEDGHSDDQKLVLAAYGTGLLTKAGLAELGIDEPLDDGPVLPRQE RIQQVLARAQ*
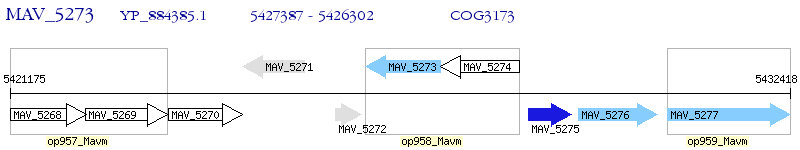
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5273 | - | - | 100% (361) | phosphotransferase enzyme family protein |
| M. avium 104 | MAV_5060 | - | 8e-26 | 27.90% (319) | putative phosphotransferase |
| M. avium 104 | MAV_0316 | - | 1e-23 | 31.23% (269) | phosphotransferase enzyme family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3193 | - | 3e-23 | 28.18% (330) | hypothetical protein Mb3193 |
| M. gilvum PYR-GCK | Mflv_1204 | - | 3e-28 | 32.57% (261) | aminoglycoside phosphotransferase |
| M. tuberculosis H37Rv | Rv3168 | - | 5e-23 | 28.18% (330) | hypothetical protein Rv3168 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4589c | - | 2e-21 | 31.20% (234) | putative phosphotransferase |
| M. marinum M | MMAR_0155 | fadE36_1 | 3e-26 | 34.40% (250) | acyl-CoA dehydrogenase FadE36_1 |
| M. smegmatis MC2 155 | MSMEG_4827 | - | 1e-24 | 31.33% (300) | aminoglycoside phosphotransferase |
| M. thermoresistible (build 8) | TH_2763 | - | 6e-29 | 31.15% (305) | PUTATIVE putative aminoglycoside phosphotransferase |
| M. ulcerans Agy99 | MUL_4942 | fadE36_1 | 2e-26 | 34.40% (250) | acyl-CoA dehydrogenase FadE36_1 |
| M. vanbaalenii PYR-1 | Mvan_5603 | - | 7e-27 | 32.21% (298) | aminoglycoside phosphotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0155|M.marinum_M --MPGPMSEDIPGVHAEAVSR--------WLVTLGHQVAAPLRFQRIG--
MUL_4942|M.ulcerans_Agy99 ------MSEDIPGVHAHAVSR--------WLVTLGHQVAAPLRFQRIG--
Mflv_1204|M.gilvum_PYR-GCK -------MTELEGLDLAALDR--------HLREVGVPRSGELRAELIA--
Mvan_5603|M.vanbaalenii_PYR-1 -------MTELEGLDLAALDR--------HLCDVGVPRSGELRAEFIS--
MAB_4589c|M.abscessus_ATCC_199 MHKDVSSDSDGPTGDAASLDADAIPTSLSEYLNRELG--EQVSVRRLS--
TH_2763|M.thermoresistible__bu ---MTAARSDTPGEELVSVAA------LDDWLDDRLPGSGPLTVERIT--
Mb3193|M.bovis_AF2122/97 -MANEPAIGAIDRLQRSSRDVTTLPAVISRWLSSVLPGGAAPEVTVESGV
Rv3168|M.tuberculosis_H37Rv -MANEPAIGAIDRLQRSSRDVTTLPAVISRWLSSVLPGGAAPEVTVESGV
MSMEG_4827|M.smegmatis_MC2_155 -----MSTFQIDTMRLADWMDG----------AALPGKGEPLTARFLS--
MAV_5273|M.avium_104 --------MTLGAERERRLTG---------WLRGRLPDADDVRVEGIDR-
MMAR_0155|M.marinum_M --LGQSNLTYRVTDAAGR-AWVLRRPPLGHLLASAHDV--------VREA
MUL_4942|M.ulcerans_Agy99 --LGQANLTYRVTDAAGR-AWVLRHPPLGHLLASAHDV--------VREA
Mflv_1204|M.gilvum_PYR-GCK --GGRSNLTFLVGDDAS--EWVLRRPPLHGLTPSAHDM--------AREY
Mvan_5603|M.vanbaalenii_PYR-1 --GGRSNLTFLVFDDAS--KWVLRRPPLHGLTPSAHDM--------AREY
MAB_4589c|M.abscessus_ATCC_199 --AGHSNLTYVVTGSSGT-QWILRRPPFGRLQAGAHDV--------MREY
TH_2763|M.thermoresistible__bu --TGHSNELFTVS-RGGP-TWLLRRPPRVKNAPTAHDM--------VREY
Mb3193|M.bovis_AF2122/97 DSTGMSSETIILTARRQQDGRSIQQKLVARVAPAAEDVPVFPTYRLDHQF
Rv3168|M.tuberculosis_H37Rv DSTGMSSETIILTARWQQDGRSIQQKLVARVAPAAEDVPVFPTYRLDHQF
MSMEG_4827|M.smegmatis_MC2_155 --GGTQNVIYELCRGDAR--CVLRMPPPGAPSDRDKGI--------LREW
MAV_5273|M.avium_104 VSFGHSAEMMTMSVIATRRGRDERRDVVLRLRPSPPALLEP--YDLARQF
* : : : ::
MMAR_0155|M.marinum_M RIMAAL-AETDVPVPRILGVTTDSEFSEAPLVLMEFVDGLVVDTMEVA--
MUL_4942|M.ulcerans_Agy99 RIMAAL-AETDVPVPRILGVTTDSEFSEAPLVLMEFVDGLVVDTMEVA--
Mflv_1204|M.gilvum_PYR-GCK KVVAAL-AGTKVPVARAVTMSDDDSVLGAPFQMVERVAGLVVRHTPQL--
Mvan_5603|M.vanbaalenii_PYR-1 RVVAAL-ADTAVPVARAVTMRNDDSVLGAPFQMVEYVAGRVVRHTDQL--
MAB_4589c|M.abscessus_ATCC_199 RVLDAL-STAQVRVPRVTLASTDAEIFGAPFYLMERQDGEVIRDVSPE--
TH_2763|M.thermoresistible__bu RVLTAL-EGTDVPHATPRGLCADPDVIGAPFYLMDKLAGVPLYHELPA--
Mb3193|M.bovis_AF2122/97 EVIRLVGELTDVPVPRVRWIETTGDVLGTPFFLMDYVEGVVPPDVMPYTF
Rv3168|M.tuberculosis_H37Rv EVIRLVGELTDVPVPRVRWIETTGDVLGTPFFLMDYVEGVVPPDVMPYTF
MSMEG_4827|M.smegmatis_MC2_155 RIIAAL-DGTDVPHTEAVAMCDDTSVLGRPFYLMGFVEGWSPMDRHGVWP
MAV_5273|M.avium_104 GILRAL-ADTDVRAPRALWLEPTGDVLGRPFFVMERVAGEVYEMQPPA--
:: : : * . .. *: :: *
MMAR_0155|M.marinum_M ----EALTPRQRRQ-IALSLPRTLAKIHAVD--LAKVGLDDLASHKPYAQ
MUL_4942|M.ulcerans_Agy99 ----EALTPRQRRQ-IALSLPRTLAKIHAVD--LAKVGLDDLASHKPYAQ
Mflv_1204|M.gilvum_PYR-GCK ----AALGDETVISNVVDSLIQVLADLHEVD--PESVGLGDFGKASGYLE
Mvan_5603|M.vanbaalenii_PYR-1 ----AALGDRATIEKCVDALIKVLADLHSVD--PDAVGLSDFGKPSGYLE
MAB_4589c|M.abscessus_ATCC_199 ----WFVGS--ARRELVLDLAVALAEIHNAD--PTRLVEAKLGRESGYLA
TH_2763|M.thermoresistible__bu ----SLAG---ERVAIAHAAIDALAALHRLD--WRAAGLDGFGKPDGYTE
Mb3193|M.bovis_AF2122/97 GDNWFADAPAERQRQLQDATVAALATLHSIPNAQNTFSFLTQGRTSDTTL
Rv3168|M.tuberculosis_H37Rv GDNWFADAPAERQRQLQDATVAALATLHSIPNAQNTFSFLTQGRTSDTTL
MSMEG_4827|M.smegmatis_MC2_155 ---EPFDSDLSTRPGLSFQLAEGIALLSKVD--WRERGLADLGRPEGFHE
MAV_5273|M.avium_104 ------DASDDTVARMCESLAEQLAAIHAVD--LTRTGLAPLDDGAGHLD
:* :
MMAR_0155|M.marinum_M RQLKRWAGQWELSKT-RELP---ELDDLTRRLVAAAPEQREI---SLVHG
MUL_4942|M.ulcerans_Agy99 RQLKRWAGQWELSKT-RELP---ELDDLTRRLVAAAPEQREI---SLVHG
Mflv_1204|M.gilvum_PYR-GCK RQVRRWGSQWDLVKR-DDDPSDADVRKLHQRLAESVPEQSRA---SIVHG
Mvan_5603|M.vanbaalenii_PYR-1 RQVRRWGSQWDLVRR-ADDPCDADVRRLHERLSAALPPQSRS---SIVHG
MAB_4589c|M.abscessus_ATCC_199 RQLKTWGGQWESIKELPTGRDLQDHTTITAWLTENYPGDRPA---AVVHG
TH_2763|M.thermoresistible__bu RQAARWGKQLKSY----AFRELPELDEVAAWLDESCPGTRGS---ALIHG
Mb3193|M.bovis_AF2122/97 HRHFNWVRSWYDFAVEGIGRS-PLLERTFEWLQSHWPDDAAAREPVLLWG
Rv3168|M.tuberculosis_H37Rv HRHFNWVRSWYDFAVEGIGRS-PLLERTFEWLQSHWPDDAAAREPVLLWG
MSMEG_4827|M.smegmatis_MC2_155 RQVDRWMAFLERIKR----RELPGLAVATDWLRAHKPLDYIP---GLMHG
MAV_5273|M.avium_104 RELGHWAAEMNRVKR----GPLPALERLHRALLSGKPAPCPR--ITLVHG
:. * * * :: *
MMAR_0155|M.marinum_M DFHLRN-LITSPETGAVVATLDWELSTLGDPLADMGSLLTYWMEPG---E
MUL_4942|M.ulcerans_Agy99 DFHLRN-LITSPETGAVVATLDWELSTLGDPLADMGSLLTYWMEPG---E
Mflv_1204|M.gilvum_PYR-GCK DYRIDNTILDADDPTKVAAVLDWEMSTLGDPLSDA-ALMCVYRLPA---F
Mvan_5603|M.vanbaalenii_PYR-1 DYRIDNTILDAEDPTKVVAVLDWEMSTLGDPLSDA-ALMCVYRHPM---F
MAB_4589c|M.abscessus_ATCC_199 DYKLDNVLVDP-ATARITAVLDWEMATVGDPLADLGYLLYMTPEPGGEVA
TH_2763|M.thermoresistible__bu DYGLHNLLYAPEPPVRILAVVDWETSTIGDPLADLGYFLASWLEPD-EVA
Mb3193|M.bovis_AF2122/97 DARVGNVLYRD---FQPVAVLDWEMVALGPRELDVAWMIFAHRVFQ----
Rv3168|M.tuberculosis_H37Rv DARVGNVLYRD---FQPVAVLDWEMVALGPRELDVAWMIFAHRVFQ----
MSMEG_4827|M.smegmatis_MC2_155 DYQFANVMYRHGAPATLAAIVDWEMGTVGDPKLDLAWMVQSWPTGA---D
MAV_5273|M.avium_104 DAKPGNFAFTG---GEVSAVFDWEMTTVGDPLTDIGWLEMLWMQPVG---
* * * .*** ::* * :
MMAR_0155|M.marinum_M SGVGDFAPSTLDGFPDRAEMTRLYLEETGRD-----PAALQYWHVLGLWK
MUL_4942|M.ulcerans_Agy99 SGVGDFAPSTLDGFPDRAEMTRLYLEETGRD-----PAALQYWHVLGLWK
Mflv_1204|M.gilvum_PYR-GCK NEVHDDAAWACDLMPPTDDLAQRYALASGKP-----LDHWDFYMALGYFK
Mvan_5603|M.vanbaalenii_PYR-1 NMVHADAAWASELIPDADALADKYSRAAGQP-----LDHWDFYMALAYFK
MAB_4589c|M.abscessus_ATCC_199 MSELTGSVTAQEGCPSHTEIAQAYREARSGDQSYWTPEDLVYYQVLGGWK
TH_2763|M.thermoresistible__bu QWGELGSPHGVAGNPSRAELIERYARKSGID---VTADDLVWYRVFGQFK
Mb3193|M.bovis_AF2122/97 ELAGLATLPGLPEVMREDDVRATYQALTGVE-----LGDLHWFYVYSGVM
Rv3168|M.tuberculosis_H37Rv ELAGLATLPGLPEVMREDDVRATYQALTGVE-----LGDLHWFYVYSGVM
MSMEG_4827|M.smegmatis_MC2_155 SGAGGMDYVDMRGMPTRDDVVAHYAEVSGRQ-----VDDLDYYLVLAKWK
MAV_5273|M.avium_104 -------INSHPAALPIDALLAHYQSASGIE-----PVNRPWYRAFNAYK
: * . :: .
MMAR_0155|M.marinum_M LAIIAEGVMRRAMDEPQNKASEGTPTVAWIDARVHMAREIADAAGMNQVS
MUL_4942|M.ulcerans_Agy99 LAIIAEGVMRRAMDEPQNKASEGTPTVAWIDARVHMAREIADAAGMNQVS
Mflv_1204|M.gilvum_PYR-GCK IAIIAAGIAYRARMGGG---------VAEGTDEVDRAVRPFVAAGLHILS
Mvan_5603|M.vanbaalenii_PYR-1 LAIIGAGIAYRAREAG----------VVDDTDKVGEAVAPLIAAGLSQLA
MAB_4589c|M.abscessus_ATCC_199 LATLLEVSYQRHLAGTT---------DDPFFAELDEGVPRLLSVARSLIP
TH_2763|M.thermoresistible__bu LAVIFEGSYGRYVRGQS---------ADPFFASLERRVPLLARHALAIAA
Mb3193|M.bovis_AF2122/97 WACVFMRTGARRVHFG-------------EIEKPDDVESLFYHAGLMKHL
Rv3168|M.tuberculosis_H37Rv WACVFMRTGARRVHFG-------------EIEKPDDVESLFYHAGLMKHL
MSMEG_4827|M.smegmatis_MC2_155 LAIVLEQGFQRAGDDEK-------------LLAFGPVVTGLMASAAELAE
MAV_5273|M.avium_104 MAVICLIGAMLVEDGHS---------DDQKLVLAAYGTGLLTKAGLAELG
* : .
MMAR_0155|M.marinum_M DSAS---------------------
MUL_4942|M.ulcerans_Agy99 DSAF---------------------
Mflv_1204|M.gilvum_PYR-GCK -------------------------
Mvan_5603|M.vanbaalenii_PYR-1 -------------------------
MAB_4589c|M.abscessus_ATCC_199 AAG----------------------
TH_2763|M.thermoresistible__bu GEA----------------------
Mb3193|M.bovis_AF2122/97 LGEEH--------------------
Rv3168|M.tuberculosis_H37Rv LGEEH--------------------
MSMEG_4827|M.smegmatis_MC2_155 STDYRG-------------------
MAV_5273|M.avium_104 IDEPLDDGPVLPRQERIQQVLARAQ