For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNRRQFLGSLAASVVAGVGAARLIVDPQPRTFAQTPAAAIPTSGPTAPQALLPPPPLSARIPLPGGGALM KIPGEGDLLALTVDDGVNSEVVRAYTQFAKDTGVRLTYFVNGVYDSWTENLQLLRPLVDSGQIQLGNHTW SHPDLTTLTKEQVAEQLSRNDAFLKKTYGIGAKPYWRPPYAKRNAAVDAVAADLGYQVPTLWSGSLSDST LITEDYILQMADQYFTPQAIVIGHLNHLPVTHVYPQLVDIIRERNLRTVTLNDVFLKTP
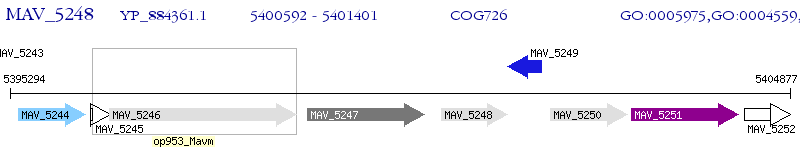
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5248 | - | - | 100% (269) | polysaccharide deacetylase domain-containing protein |
| M. avium 104 | MAV_1218 | - | 2e-13 | 29.38% (194) | carbohydrate degrading enzyme |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1126 | - | 2e-11 | 28.12% (192) | glycosyl hydrolase |
| M. gilvum PYR-GCK | Mflv_2063 | - | 4e-08 | 28.80% (191) | polysaccharide deacetylase |
| M. tuberculosis H37Rv | Rv1096 | - | 2e-11 | 28.12% (192) | glycosyl hydrolase |
| M. leprae Br4923 | MLBr_02649 | - | 1e-123 | 77.61% (268) | hypothetical protein MLBr_02649 |
| M. abscessus ATCC 19977 | MAB_2590 | - | 2e-80 | 56.55% (267) | putative polysaccharide deacetylase |
| M. marinum M | MMAR_4371 | - | 1e-12 | 28.65% (192) | glycosyl hydrolase |
| M. smegmatis MC2 155 | MSMEG_5023 | - | 6e-09 | 22.28% (202) | chitooligosaccharide deacetylase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0184 | - | 3e-13 | 29.17% (192) | glycosyl hydrolase |
| M. vanbaalenii PYR-1 | Mvan_4651 | - | 9e-12 | 28.50% (193) | polysaccharide deacetylase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4371|M.marinum_M --MRKRPDNQAWRYWRMVVGVVAAVAVLV---------------------
MUL_0184|M.ulcerans_Agy99 --MRKRPDDQAWRYWRMVVGVVAAVAVLV---------------------
Mb1126|M.bovis_AF2122/97 --MPKRPDNQTWRYWRTVTGVVVAGAVLV---------------------
Rv1096|M.tuberculosis_H37Rv --MPKRPDNQTWRYWRTVTGVVVAGAVLV---------------------
Mflv_2063|M.gilvum_PYR-GCK --MTSESGALDDAHRRQARPIRRLGAVVLG--------------------
Mvan_4651|M.vanbaalenii_PYR-1 --MTSGSGAQYDTRGARARRIR-SAAILLG--------------------
MSMEG_5023|M.smegmatis_MC2_155 ---MRRRVLTVCLALVLCVVVVLAGGYWLS--------------------
MAV_5248|M.avium_104 ---MNRRQFLGSLAASVVAGVGAARLIVDPQPRTFAQTPAAAIPTSGPTA
MLBr_02649|M.leprae_Br4923 MSALNRRSFLGALAVSTVTGLGAVRLVVGPQPRTFVQAPVVAELTPTPNA
MAB_2590|M.abscessus_ATCC_1997 --MFDRRRFLTLLAGAAAAGASFIEYVS-----------AAADPGTAAGS
MMAR_4371|M.marinum_M ----------IGSLTGHV---TRADDQSCAVVKCVALTFDDGP-SPFTDR
MUL_0184|M.ulcerans_Agy99 ----------IGSLTGHV---TRADDQSCAVVKCVALTFDDGP-SPFTDR
Mb1126|M.bovis_AF2122/97 ----------VGGLSGRV---TRAENLSCSVIKCVALTFDDGP-GPYTDR
Rv1096|M.tuberculosis_H37Rv ----------VGGLSGRV---TRAENLSCSVIKCVALTFDDGP-GPYTDR
Mflv_2063|M.gilvum_PYR-GCK ---------VITVFTAAP--TAAAGPVDCAAVPCVALTFDDGP-GPHTDR
Mvan_4651|M.vanbaalenii_PYR-1 ---------LVGGFIAAVPAPAAADSVDCARVKCVALTFDDGP-GPFTDR
MSMEG_5023|M.smegmatis_MC2_155 ---------NSRTFQVAG----ALVHRVSTPEKVVALTLDDGP-TDATRV
MAV_5248|M.avium_104 PQALLPPPPLSARIPLPGGGALMKIPGEGD---LLALTVDDGVNSEVVRA
MLBr_02649|M.leprae_Br4923 -AGVLPPPPTSARIPLPGGGVLSKIPGRGD---LLALTVDDGVNTEVVRS
MAB_2590|M.abscessus_ATCC_1997 SNSGLP------RVPLPGGGTLTALPSRTDGQRLLALTLDDGTDANVIAA
. :***.***
MMAR_4371|M.marinum_M LLQILKDNDAKATFFLIG--NKVAANPAAAKRIAEAG-MEIGSHTWEHPN
MUL_0184|M.ulcerans_Agy99 LLQILKDNDAKATFFLIG--NKVAANPAAAKRIAEAG-MEIGSHTWEHPN
Mb1126|M.bovis_AF2122/97 LLHILTDNDAKATFFLIG--NKVAANPAGARRIADAG-MEIGSHTWEHPN
Rv1096|M.tuberculosis_H37Rv LLHILTDNDAKATFFLIG--NKVAANPAGARRIADAG-MEIGSHTWEHPN
Mflv_2063|M.gilvum_PYR-GCK LLDVLRAQGAKATFFVIG--EKVAADPAATRRITGAG-MQVGNHTWQHLD
Mvan_4651|M.vanbaalenii_PYR-1 LLQTLRANNAEATFFLIG--DKVAADPAAARRIVDAG-MEIGNHTWSHPD
MSMEG_5023|M.smegmatis_MC2_155 VLRTLAAARVPATFYLTG--RELEAAPDLGAAIAAAG-HEIGNHSFSHRR
MAV_5248|M.avium_104 YTQFAKDTGVRLTYFVNGVYDSWTENLQLLRPLVDSGQIQLGNHTWSHPD
MLBr_02649|M.leprae_Br4923 YVQFVKDTDIRLTFFVNGVYDSWTDNMSMLRPLVESGQIQLGNHTWSHPD
MAB_2590|M.abscessus_ATCC_1997 YTQLARDTGMRLTYFVNGVNAGWTQNAPMLRPLVDSGQIQLGNHTWSHPN
*::: * :. :* ::*.*::.*
MMAR_4371|M.marinum_M MTTIPEADIAGQFSRANDAIKAATGRTP-TLYRPAGGLSNAAVRQTAEKF
MUL_0184|M.ulcerans_Agy99 MTTIPEADIAGQFSRANDAIKAATGRTP-TLYRPAGGLSNAAVRQTAAKY
Mb1126|M.bovis_AF2122/97 MTTIPPEDIPGQFSRANDVIAAATGRTP-TLYRPAGGLSNDAVRQAAAKV
Rv1096|M.tuberculosis_H37Rv MTTIPPEDIPGQFSRANDVIAAATGRTP-TLYRPAGGLSNDAVRQAAAKV
Mflv_2063|M.gilvum_PYR-GCK MTTLAPPDVAAQFARATEAIAAATGQRT-PLARTGFGAIDDSVLAEAGRQ
Mvan_4651|M.vanbaalenii_PYR-1 LTAIPPQELPAQLSRATEVLERATGERP-TLMRPPFGAVDDAVLAAAGSQ
MSMEG_5023|M.smegmatis_MC2_155 MVLMSSKTVASELERTDAAIRATGYTGP-ITFRPPYGKKLWSLPRYLAEH
MAV_5248|M.avium_104 LTTLTKEQVAEQLSRNDAFLKKTYGIGAKPYWRPPYAKRNAAVDAVAADL
MLBr_02649|M.leprae_Br4923 LTAMTKSEIATQLNRNDDFLKKNYGIAAQPYWRPPYAKHNATVDAVAVDL
MAB_2590|M.abscessus_ATCC_1997 LTTLTAEQIADQMARNTKFLVNTYGVDPAPYFRPPYGKHNAATDSVTRGL
:. :. :. :: * : . *. . :
MMAR_4371|M.marinum_M GQAEILWDVIPFDWANDSNTAATRYMLMRQIKPGSVVLFHDTYSS---TV
MUL_0184|M.ulcerans_Agy99 GQAEILWDVIPFDWANDSNTAATRYMLMRQIKPGSVVLFHDTYSS---TV
Mb1126|M.bovis_AF2122/97 GQAEILWDVIPFDWINDSNTAATRHMLMTQIKPGSVVLFHDTYSS---TV
Rv1096|M.tuberculosis_H37Rv GQAEILWDVIPFDWINDSNTAATRHMLMTQIKPGSVVLFHDTYSS---TV
Mflv_2063|M.gilvum_PYR-GCK GLTAVNWDVNPRDWHHDADPDAIRDAVLTQVRPGAVVLLHDTVAA---TV
Mvan_4651|M.vanbaalenii_PYR-1 GLAVVNWDVIPLDWENDTDIAATRAALTEQIRPNSVVLLHDTFAS---TV
MSMEG_5023|M.smegmatis_MC2_155 DRISVTWDVEP-DSATEPTADEIVAQTVDQVRPGSIILLHAMYGSRGPTR
MAV_5248|M.avium_104 GYQVPTLWSGSLSDSTLITEDYILQMADQYFTPQAIVIGHLNHLP---VT
MLBr_02649|M.leprae_Br4923 GYTVPTLWSGSLSDSTLITEDYIVKMADQYFTPQAIVIGHLNHLP---VT
MAB_2590|M.abscessus_ATCC_1997 GYGPPVLWYGSLSDSGVVTPEYIVDMARKYFNPQAIVIGHLNHPP---VT
. . . . * :::: * . .
MMAR_4371|M.marinum_M DLVYQFIPVLKANGYRLVTVSELLGPRAPGSSYGSRDNGPPVDELRDIPP
MUL_0184|M.ulcerans_Agy99 DLVYQFIPVLKANGYRLVTVSELLGPRAPGNSYGSRDNGPPVDELRDIPP
Mb1126|M.bovis_AF2122/97 DVVYQFIPVLKANGYRLVTVSELLGPRAPGSSYGSRENGPPVNELRDIPA
Rv1096|M.tuberculosis_H37Rv DVVYQFIPVLKANGYRLVTVSELLGPRAPGSSYGSRENGPPVNELRDIPA
Mflv_2063|M.gilvum_PYR-GCK GAMADVVPALRARGYHLVTVSQLLGPLTPGSLYGSRERA-----------
Mvan_4651|M.vanbaalenii_PYR-1 DLMAEFVPVLRANGYHLVTVSQMIGPRPPGSLYGTPP-------------
MSMEG_5023|M.smegmatis_MC2_155 AALPRIISELQSAGYRFVTVSQLIGMS-----------------------
MAV_5248|M.avium_104 HVYPQLVDIIRERNLRTVTLNDVFLKTP----------------------
MLBr_02649|M.leprae_Br4923 HVYPQLIDIIRSRHLRTVTLNDVFLTTS----------------------
MAB_2590|M.abscessus_ATCC_1997 TVYRQLLDVIGERQLRTVTLNDVYQH------------------------
.: : : **:.::
MMAR_4371|M.marinum_M SDIPPLPNTPSPKPMPNFPITDIAGQNSGGPNNGA
MUL_0184|M.ulcerans_Agy99 SDIPPLPNTPSPKPMPNFPITDIAGQNSGGPNNGA
Mb1126|M.bovis_AF2122/97 SEIPPLPNTSSPKPMSNFPITDIAGQNSGGPNNGA
Rv1096|M.tuberculosis_H37Rv SEIPPLPNTSSPKPMPNFPITDIAGQNSGGPNNGA
Mflv_2063|M.gilvum_PYR-GCK -----------------------------------
Mvan_4651|M.vanbaalenii_PYR-1 -----------------------------------
MSMEG_5023|M.smegmatis_MC2_155 -----------------------------------
MAV_5248|M.avium_104 -----------------------------------
MLBr_02649|M.leprae_Br4923 -----------------------------------
MAB_2590|M.abscessus_ATCC_1997 -----------------------------------