For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VWDFETDPEYQAKLDWVEKFMAEELEPLDLVALDPYDKKNAEMMAILRPLQQQVKDQGLWAAHLRPELGG QGFGQVKLALLNEILGRSRWAPSVFGCQAPDSGNAEILALFGTDEQKARYLQPLLDGEITSCYSMTEPQG GSDPGQFVTAATRDGDYWVINGEKWFSTNAKHASFFIVMAVTNPEARTYDKMSLFIVPAETPGIEIVRNV GVGAESSSRASHGYVGYTDVRVPADHVLGGEGQAFMIAQTRLGGGRIHHAMRTIALARSAFDMMCERAVS RKTRHGRLADFQMTQEKIADSWIQIEQFRLLVLRTAWLIDKHHDYQKVRRDIAAVKVAMPQVLHDVAQRA LHLHGALGVSDEMPFVKMLVAAESLGIADGATELHKMTVARRTLREYEPVSTPFPSAHIPTRRAEAQARL AERLEHAVAEF
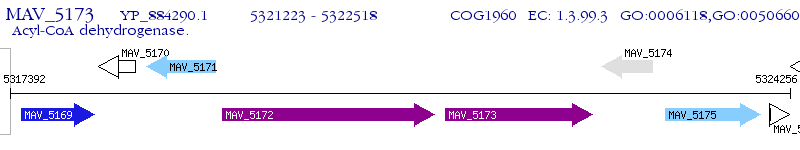
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5173 | - | - | 100% (431) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_3944 | - | e-147 | 59.34% (423) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_5144 | - | 3e-63 | 38.96% (367) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0136c | fadE1 | 0.0 | 90.57% (435) | acyl-CoA dehydrogenase FADE1 |
| M. gilvum PYR-GCK | Mflv_2451 | - | 1e-153 | 60.42% (432) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0131c | fadE1 | 0.0 | 90.57% (435) | acyl-CoA dehydrogenase FADE1 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 1e-25 | 25.81% (341) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4911c | - | 1e-165 | 63.70% (427) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_0331 | fadE1 | 0.0 | 92.34% (431) | acyl-CoA dehydrogenase FadE1 |
| M. smegmatis MC2 155 | MSMEG_4826 | - | 1e-156 | 62.15% (428) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2939 | - | 1e-152 | 61.15% (435) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4790 | fadE1 | 0.0 | 91.42% (431) | acyl-CoA dehydrogenase FadE1 |
| M. vanbaalenii PYR-1 | Mvan_4200 | - | 1e-152 | 60.14% (429) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2451|M.gilvum_PYR-GCK -------------MWSFETDPEYQKV-LDWADEFVREEVEPLDLAFPHQQ
Mvan_4200|M.vanbaalenii_PYR-1 -------------MWSFETDPEYQKV-LDWADEFVREEVEPLDLAFPHQQ
TH_2939|M.thermoresistible__bu ------------MAWDFETDPEYQKV-LDWADEFVRTEVEPLDLAFPHQQ
MSMEG_4826|M.smegmatis_MC2_155 ------------MAWDFETDPQYQEV-LDWADEFVREKVEPLDLVWPHLQ
Mb0136c|M.bovis_AF2122/97 MPVRRRAGERLPTVWDFETDPQYQSK-LDWVEKFMAEELEPLDLVALDP-
Rv0131c|M.tuberculosis_H37Rv MPVRRRAGERLPTVWDFETDPQYQSK-LDWVEKFMAEELEPLDLVALDP-
MMAR_0331|M.marinum_M -------------MWDFETDPEYQAK-LDWVQEFMVNELEPLDLVALDP-
MUL_4790|M.ulcerans_Agy99 -------------MWDFETDPEYQAK-LDWVQEFMVNELEPLDLVALDP-
MAV_5173|M.avium_104 -------------MWDFETDPEYQAK-LDWVEKFMAEELEPLDLVALDP-
MAB_4911c|M.abscessus_ATCC_199 ---------MTAMAWDFETDPEYQEL-LDWADEFVRTEVEPLDYVFPNP-
MLBr_00737|M.leprae_Br4923 -------------MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAP-
.:. :* :: * .: : :..*
Mflv_2451|M.gilvum_PYR-GCK FVPLDGKRREAIDPLKEEVRRRGLWATHLGADLGGQGYGQLKLALLNEIL
Mvan_4200|M.vanbaalenii_PYR-1 FVPLDGERRRAIDPLKEEVRRRGLWATHLGADLGGQGYGQLKLALLNEIL
TH_2939|M.thermoresistible__bu FVPLEGTRREAIEPLKQEVRRRGLWATHLGPELGGQGYGQLKLALLNEIL
MSMEG_4826|M.smegmatis_MC2_155 FTPLNEARRQVVDPLKEQVRRKGLWATHLGPELGGQGYGQLKLALLNEIL
Mb0136c|M.bovis_AF2122/97 YDKKNADTMAILRPLQRQVKDQGLWAAHLRPELGGQGFGQVKLALLNEII
Rv0131c|M.tuberculosis_H37Rv YDKKNADTMAILRPLQRQVKDQGLWAAHLRPELGGQGFGQVKLALLNEII
MMAR_0331|M.marinum_M YDKKNPEMMAVLRPLQQQVKDQGLWAAHLRPELGGQGFGQVKLALLNEIL
MUL_4790|M.ulcerans_Agy99 YDKKNPEMMAVLRPLQQ-VKNQGLWAAHLRPEVGGQGFGQVKLALLNEIL
MAV_5173|M.avium_104 YDKKNAEMMAILRPLQQQVKDQGLWAAHLRPELGGQGFGQVKLALLNEIL
MAB_4911c|M.abscessus_ATCC_199 YDKSDTVAMDYVRPLKEEVKRRGLWACHLGPELGGPGYGQLKLALLNEIL
MLBr_00737|M.leprae_Br4923 HAADVDQRARFPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEV
. :. *: * *: : ** * ..: .:: * :
Mflv_2451|M.gilvum_PYR-GCK GRSQWAPIIFGCQAPDTGNAEIIAHYGTEEQKQRYLRPLLDGELFSCYSM
Mvan_4200|M.vanbaalenii_PYR-1 GRSQWAPVIFGCQAPDTGNAEIIAHYGTEEQKQKYLRPLLDGELFSCYSM
TH_2939|M.thermoresistible__bu GRSQWAPVVFGCQAPDTGNAEIIAHYGTEEQQKRYLQPLLDGELFSCYSM
MSMEG_4826|M.smegmatis_MC2_155 GRSQWAPIVFGCQAPDTGNAEIIAHYGTDEQKERYLRPLLEGELFSSYSM
Mb0136c|M.bovis_AF2122/97 GRSRWAPSAFGCQAPDSGNAEILALFGTDEQKARYLRPLLDGEITSCYSM
Rv0131c|M.tuberculosis_H37Rv GRSRWAPSAFGCQAPDSGNAEILALFGTDEQKARYLRPLLDGEITSCYSM
MMAR_0331|M.marinum_M GRSRWAPSVFGCQAPDSGNAEILALFGTEDQKARYLQPLLDGEITSCYSM
MUL_4790|M.ulcerans_Agy99 GRSRWAPSVFGCQAPDSGNAEILALFGTEDQKARYLQPLLDGEITSCYSM
MAV_5173|M.avium_104 GRSRWAPSVFGCQAPDSGNAEILALFGTDEQKARYLQPLLDGEITSCYSM
MAB_4911c|M.abscessus_ATCC_199 GRSVWAPSIFGCQAPDTGNAEILAHYGTEAQKAKYLQPLMDGDIGSCYVM
MLBr_00737|M.leprae_Br4923 ARVDASASLIPAVN-KLGTMGLILR-GSEELKKQVLPSLAAEGAMASYAL
.* :. : . . *. :: *:: : : * .* :.* :
Mflv_2451|M.gilvum_PYR-GCK TEPHAGADPTMFTTSAVRDGD----DWVINGWKFFSSNAATASFLIVMVV
Mvan_4200|M.vanbaalenii_PYR-1 TEPHAGADPTMFTTSAVRDGD----DWVINGWKFFSSNAATASFLIVMVV
TH_2939|M.thermoresistible__bu TEPHAGADPTLFKTTAVRDGAGPDADWVINGWKFFSSNATTASFLIVMAV
MSMEG_4826|M.smegmatis_MC2_155 TEPHAGADPTLFTTRAVRDGD----DWVINGWKFFSSNAATASFLIVMVV
Mb0136c|M.bovis_AF2122/97 TEPQGGSDPGLFVTAATRDAAG-NGDWIINGEKWFSTNAKHASFFIVMAV
Rv0131c|M.tuberculosis_H37Rv TEPQGGSDPGLFVTAATRDAAG-NGDWIINGEKWFSTNAKHASFFIVMAV
MMAR_0331|M.marinum_M TEPQGGSDPGSFVTAATRDGD----SWIINGEKWFSTNAKHASFFIVMAV
MUL_4790|M.ulcerans_Agy99 TEPQGGSDPGSFVTAATRDGD----SWIINGEKWFSTNAKHASFFIVMAV
MAV_5173|M.avium_104 TEPQGGSDPGQFVTAATRDGD----YWVINGEKWFSTNAKHASFFIVMAV
MAB_4911c|M.abscessus_ATCC_199 TEPQGGSDPTLFKTTAVRDGD----EWILNGEKWFNSEARHAAFWIVMAV
MLBr_00737|M.leprae_Br4923 SEREAGSDAASMRTRAKADGD----DWILNGFKCWITNGGKSTWYTVMAV
:* ..*:*. : * * *. *::** * : ::. ::: **.*
Mflv_2451|M.gilvum_PYR-GCK TNPDVSAYQGMSMFLVPTDTPGVTIERNVGLYGEREGEGSHALIHYDNVR
Mvan_4200|M.vanbaalenii_PYR-1 TNREVSAYQGMSMFLVPTDTPGVKIERNIGLYGEGAEHGSHALIHYDNVR
TH_2939|M.thermoresistible__bu TNPDVSAYQGMSMFLVPTDTPGVKIERSIGLYGDPPGTGSHALIHYDNVR
MSMEG_4826|M.smegmatis_MC2_155 TNPDVSAYQGMSMFLVPTDTPGVEIVRNVGLYGERDNEGSHALIHYDNVR
Mb0136c|M.bovis_AF2122/97 TKPEARTYEKMSLFIVPADTPGIEIVRNVGVGAESTRHASHGYIRYHDVR
Rv0131c|M.tuberculosis_H37Rv TKPEARTYEKMSLFIVPADTPGIEIVRNVGVGAESTRHASHGYIRYHDVR
MMAR_0331|M.marinum_M TNPQARTYEKMSLFIVPAETPGIEIVRNVGVGTESSKHASHGYVRYHDVR
MUL_4790|M.ulcerans_Agy99 TNPQARTYEKMSLFIVPAETPGIEIVRNVGVGTESSKHASHGYVRYHDVR
MAV_5173|M.avium_104 TNPEARTYDKMSLFIVPAETPGIEIVRNVGVGAESSSRASHGYVGYTDVR
MAB_4911c|M.abscessus_ATCC_199 TNTEVSPYQGMSMFIVDPNAPGVEIVRNITVHGFS---EDEAYTRFTNAR
MLBr_00737|M.leprae_Br4923 TDPDK-GANGISAFIVHKDDEGFSIGPKE--KKLGIKGSPTTELYFDKCR
*. : : :* *:* : *. * . : . *
Mflv_2451|M.gilvum_PYR-GCK VPSEALLGGEGQAFAIAQTRLGGGRIHHAMRTIGLSRKALDMMCERALSR
Mvan_4200|M.vanbaalenii_PYR-1 VPAEALLGGEGQAFVIAQTRLGGGRIHHAMRTIGLSRKALDMMCERALSR
TH_2939|M.thermoresistible__bu VPADALLGGEGQAFAIAQTRLGGGRIHHAMRTIGLARKALDMMCERALSR
MSMEG_4826|M.smegmatis_MC2_155 VPAEALLGGEGQAFVIAQTRLGGGRIHHAMRTIGLARKALDMMCERALSR
Mb0136c|M.bovis_AF2122/97 VPADHVLGGEGQAFMIAQTRLGGGRIHHAMRTIALARRAFDMMCERALSR
Rv0131c|M.tuberculosis_H37Rv VPADHVLGGEGQAFMIAQTRLGGGRIHHAMRTIALARRAFDMMCERALSR
MMAR_0331|M.marinum_M VPADHVLGGEGQAFMIAQTRLGGGRIHHAMRTIALARRAFDMMCERAVSR
MUL_4790|M.ulcerans_Agy99 VPADHVLGGEGQAFMTAQTRLGGGRIHHAMRTIALARRAFDMMCERAVSR
MAV_5173|M.avium_104 VPADHVLGGEGQAFMIAQTRLGGGRIHHAMRTIALARSAFDMMCERAVSR
MAB_4911c|M.abscessus_ATCC_199 IPAENMLGAEGGAFAIAQTRLGGGRVHHAMRTVAQVRKAFDMMCERVVSR
MLBr_00737|M.leprae_Br4923 IPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDR
:*.: ::* * .* * : *. * . :::. : *:* . .*
Mflv_2451|M.gilvum_PYR-GCK ETQGSRLSDKQFVQGYIADSYAQLLQFRLMVLYTAWEIDKYNDYKKVRKD
Mvan_4200|M.vanbaalenii_PYR-1 ETQGSRLSDKQFVQGYIADSYAQLLQFRLMVLYTAWEIDKYNDYKKVRKD
TH_2939|M.thermoresistible__bu QTQGSRLSDKQFVQGYIADSYAQLLQFRLMVLYTAWEIDKYNDYKKVRKD
MSMEG_4826|M.smegmatis_MC2_155 KTQGSRLSDKQFVQGYIADSYAQLLQFRLMVLYTAWEIDKYNDYKLVRKD
Mb0136c|M.bovis_AF2122/97 QTRHGRLADLQMTQEKIADSWIQIEQFRLLVLRTAWLIDKHHDYQKVRRD
Rv0131c|M.tuberculosis_H37Rv QTRHGRLADLQMTQEKIADSWIQIEQFRLLVLRTAWLIDKHHDYQKVRRD
MMAR_0331|M.marinum_M QTRHGRLSDLQMTQEKVADSWIQIEQFRLLVLRTAWLIDKHHDYQKVRRD
MUL_4790|M.ulcerans_Agy99 QTRHGRLSDLQMTQEKVADSWIQIEQFRLLVLRTAWLIDKHHDYQKVRRD
MAV_5173|M.avium_104 KTRHGRLADFQMTQEKIADSWIQIEQFRLLVLRTAWLIDKHHDYQKVRRD
MAB_4911c|M.abscessus_ATCC_199 EARGGTIAKLQMTQERIADSWIEMESFRLLVLRTAWLIDKHQDYLKVRKD
MLBr_00737|M.leprae_Br4923 KQFGESISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFIS-
: :: * * :** :: **:* :* :: . .
Mflv_2451|M.gilvum_PYR-GCK IAAVKVVMPTVLHDIAWRAMQVHGALGVTNEVPFLGMVTGAAVMGLADGP
Mvan_4200|M.vanbaalenii_PYR-1 IAAVKVVMPTVLHDIAWRAMQVHGALGVTNEVPFLGMVTGAAVMGLADGP
TH_2939|M.thermoresistible__bu IAAVKATMPTVLHDIAWRAMQVHGALGVTDEMPFMGMITGAAVMGLADGP
MSMEG_4826|M.smegmatis_MC2_155 IAAVKVVMPTVLHDIAWRAMQVHGALGVTNEMPFLGMVTGAAVMGLADGP
Mb0136c|M.bovis_AF2122/97 IAAVKVAMPQVLHDVVQRAMHLHGALGVSDEMPFVKMMLAAESLGIADGA
Rv0131c|M.tuberculosis_H37Rv IAAVKVAMPQVLHDVVQRAMHLHGALGVSDEMPFVKMMLAAESLGIADGA
MMAR_0331|M.marinum_M IAAVKVAMPQVLHDVVQRAMHLHGALGVSDEMPFVKMMVAAESLGIADGA
MUL_4790|M.ulcerans_Agy99 IAAVKVAMPQVLHDVVQRAMHLHGALGVSDEMPFVKMMVAAESLGIADGA
MAV_5173|M.avium_104 IAAVKVAMPQVLHDVAQRALHLHGALGVSDEMPFVKMLVAAESLGIADGA
MAB_4911c|M.abscessus_ATCC_199 ISAIKAAMPKVMHDVAQRALHLHGSLGVSEEMPLAKMFLYSEVMGLVDGP
MLBr_00737|M.leprae_Br4923 -AASKCFASDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGT
:* * . : ::. *:::.*. * :.:.*. :. : : :*.
Mflv_2451|M.gilvum_PYR-GCK TEVHKTTVAKQVLRDHRPAEGMWPSEWIPAKREAAQAKYADFIEHEVGNL
Mvan_4200|M.vanbaalenii_PYR-1 TEVHKTTVAKQVLRDYRPAEGLWPSEWIPAKREAARAKYAEYLENEVGNL
TH_2939|M.thermoresistible__bu TEVHKATVAKQVLRDYRPSDDVWPSEWIPRKREAALAKYAEYLEHEVGNL
MSMEG_4826|M.smegmatis_MC2_155 TEVHKTTVAKQVLRDYQPTEDTWPTEWIPRKREAARAKLADYLENEVGNL
Mb0136c|M.bovis_AF2122/97 TELHKMTVARRTLREYQPVTTLFPSQHIPTRRAHAEAWLAQRLEHAIAEF
Rv0131c|M.tuberculosis_H37Rv TELHKMTVARRTLREYQPVTTLFPSQHIPTRRAHAEAWLAQRLEHAIAEF
MMAR_0331|M.marinum_M TELHKMTVARRTLRQYEPVTTPFPSQHIPTRRAAAEARLAERLEHSIAEF
MUL_4790|M.ulcerans_Agy99 TELHKMTVARRTLRQYEPVATPFPSQHIPTRRAAAEARLAERLEHSIAEF
MAV_5173|M.avium_104 TELHKMTVARRTLREYEPVSTPFPSAHIPTRRAEAQARLAERLEHAVAEF
MAB_4911c|M.abscessus_ATCC_199 TEVHKVTIAKEVLKDYTPYEGLFPPSHIPALTEKARAHVAARLEHRVGNL
MLBr_00737|M.leprae_Br4923 NQIQRVVMSRALLR------------------------------------
.:::: .::: *: