For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAKSIGETPRRSGVKSPPTARVMDVLAAVADSPGGLTSAELAKGCAISSSTCALVLAELERRAWVTRRDD RRYVLGSGLFGLVHGLREQFPLLDRGRDALRLLHERLGAGCSMSKIGARHLTTVDSVGHGTDGEHAVGQR FPIDPPFGLVAMAWRDDDAVQAWLRRVTPRLTRTEIAQHQRVLADIRARGYGAWRFDDTHRSLHNRLAEA LASLEPTAQVARRLTTLMTMVTLRSVTDVLETELPTTEFVVLPIFGHDGQPEYQIEIHLGHSAGLTLPEL DDALEQARRLLTAPVR
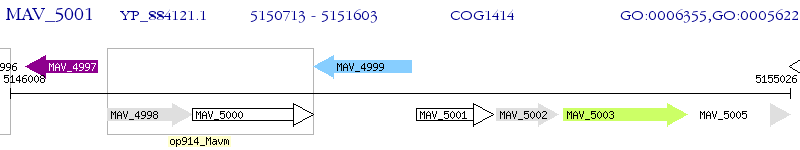
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5001 | - | - | 100% (296) | hypothetical protein MAV_5001 |
| M. avium 104 | MAV_3545 | - | 4e-15 | 29.28% (222) | hypothetical protein MAV_3545 |
| M. avium 104 | MAV_5235 | - | 8e-12 | 24.34% (267) | putative transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1528 | - | 5e-21 | 30.00% (290) | regulatory protein, IclR |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3479c | - | 6e-20 | 27.74% (274) | IclR family regulatory protein |
| M. marinum M | MMAR_3507 | - | 3e-12 | 29.80% (255) | IclR family transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5933 | - | 1e-19 | 27.61% (268) | hypothetical protein MSMEG_5933 |
| M. thermoresistible (build 8) | TH_0373 | - | 4e-21 | 30.42% (263) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2762 | - | 8e-13 | 30.20% (255) | IclR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5230 | - | 1e-17 | 29.30% (273) | regulatory proteins, IclR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5933|M.smegmatis_MC2_155 ------------------------MLEQTDAGRAS-----PPTARVVAIL
TH_0373|M.thermoresistible__bu ------------------------MVETGPVGRTS-----PPTERVVQVL
Mflv_1528|M.gilvum_PYR-GCK ------------------------MSVPVQPGRAS-----PPTDRVVQIL
Mvan_5230|M.vanbaalenii_PYR-1 ------------------------MSEPPAPGRAS-----PPTDRVVRVL
MMAR_3507|M.marinum_M ------------------------MTDTVDPASVSSQGSSPPTERVIAVL
MUL_2762|M.ulcerans_Agy99 MPSPPALASVEISTYRSIDIPFDILTGTVDPASVSSQGLSPPTERVIAVL
MAV_5001|M.avium_104 ------------------------MAKSIGETPRRSGVKSPPTARVMDVL
MAB_3479c|M.abscessus_ATCC_199 ------------------------MTKP-----------APAVVRAAQIL
: *.. *. :*
MSMEG_5933|M.smegmatis_MC2_155 DFLARHPHDRFGLSELSRRVGVSKPTCLGILTTLTESGYLVREGDGRDKC
TH_0373|M.thermoresistible__bu ELLARHPHERFGASEVARRLNLSKPTCLGILATLARHGYAVREP--TDKT
Mflv_1528|M.gilvum_PYR-GCK DFLASRPTERYGVSDLARRVGLSKPTCLGIVTSLTEAGYLVRDP--ADKT
Mvan_5230|M.vanbaalenii_PYR-1 DFLARHPQERFGVSELARRVDLTKPTCLGIVTSLRDAGYLVRDP--ADKT
MMAR_3507|M.marinum_M ELLGHNSAKQFSLAEICRGLGISRATGHAILTTLAAREWVIRDP--ETFR
MUL_2762|M.ulcerans_Agy99 ELLGHNSAKQFSLAEICRGLGISRATGHAILTTLAAREWVIRDP--ETFR
MAV_5001|M.avium_104 AAVADSPGG-LTSAELAKGCAISSSTCALVLAELERRAWVTRRD---DRR
MAB_3479c|M.abscessus_ATCC_199 DHLAGRPTLPLSMTEIAEAVGVNPASTLAILQALTEAGYIVRHP--RHKT
:. . :::.. :. .: :: * : *
MSMEG_5933|M.smegmatis_MC2_155 YRLGPALIALGHMAQESMRVNPFARAELRALSGAFDSTAAISAVIDDRIT
TH_0373|M.thermoresistible__bu YRPGPALIALGHAAQESLRVSPDARAQLRELSGAFGGTAALSAVVDDRIT
Mflv_1528|M.gilvum_PYR-GCK YRLGPSLITLGHRAQESMRVSPAARDHLRRLSARYGATAALSAVVDDRIT
Mvan_5230|M.vanbaalenii_PYR-1 YRLGPSLITLGHKAQESMRVSPAAREQLRRLSTRFGATAALSAVVDDRIT
MMAR_3507|M.marinum_M YSWGPAIAGLDTPTTAQLHRG-----DLQELAAATGTQVMLARREDATLV
MUL_2762|M.ulcerans_Agy99 YSWGPAIAGLDTPTTAQLHRG-----DLQELAAATGTQVMLARREDATLV
MAV_5001|M.avium_104 YVLGSGLFGLVHGLREQFPLLDRGRDALRLLHERLGAGCSMSKIGARHLT
MAB_3479c|M.abscessus_ATCC_199 YSIGPAMLITGHAARMRFEVADVVNAEVQRLASQFGTACTASMIAGEDVA
* *..: : :: * . : :.
MSMEG_5933|M.smegmatis_MC2_155 VLELVAPPGTDVGVRVGQSYPFAPPVGLMYVLWDD-DALRAWLDKSPTVP
TH_0373|M.thermoresistible__bu VLELVAPPGATPPVHVGQSYPFAPPVGLMFVLWDD-EAVTAWLAKEPTIP
Mflv_1528|M.gilvum_PYR-GCK LLDLVAPTGARPGVEVGQSYPFAPPVGLMFVLWDD-EAEQDWLRRQPTIP
Mvan_5230|M.vanbaalenii_PYR-1 LLDLVAPVGARPGVEVGQSYPFAPPVGLMFVLWDD-EAVRDWLAMEPTVP
MMAR_3507|M.marinum_M VIDTVGECPRGPRISRGMRTPFVAPIGRDYVAWWSPDAQQDWLRAFGAPN
MUL_2762|M.ulcerans_Agy99 VIDTVGECPRGPRISRGMRTPFVAPIGRDYVAWWSPDAQQDWLRAFGAPN
MAV_5001|M.avium_104 TVDSVG-HGTDGEHAVGQRFPIDPPFGLVAMAWRDDDAVQAWLRRVTPRL
MAB_3479c|M.abscessus_ATCC_199 VIASHGRSPTGASYQVGQRIRLSAPAGLVFMAWAPDDGVRRWLLRATPVI
: . * : .* * : * :. ** .
MSMEG_5933|M.smegmatis_MC2_155 LR---TDSERLEKVIGECRNSGYLVERLTPAGQRLYGLMAGMSSR-LPDE
TH_0373|M.thermoresistible__bu LR---TDSERLQRVIAECRADGYLVERQTPAGQRLYTLMAGMSAT-LPEE
Mflv_1528|M.gilvum_PYR-GCK LR---TDTERLKRVVASCRADGYLVERQTPGGRRLYALMAGMPTE-LPDE
Mvan_5230|M.vanbaalenii_PYR-1 LHPSRTDPDRLHRVIRRCRADGYLVERLTPGGRRLYSLMAGMSST-LPDE
MMAR_3507|M.marinum_M PR----FLQRMTLVLNQIQRRGFVVERLTRQYVRVYTALRALSADGEVDE
MUL_2762|M.ulcerans_Agy99 PR----FLQRMILVLNQIQRRGFVVERLTRQYVRVYTALRALSADGEVDE
MAV_5001|M.avium_104 TR---TEIAQHQRVLADIRARGYGAWRFDDTHRSLHNRLAEALAS---LE
MAB_3479c|M.abscessus_ATCC_199 TD---ELWDGMQAMLADIRRQGYFASVQSDRVDKFYMQLGDTAAHGTVHE
:: : *: . .: : : *
MSMEG_5933|M.smegmatis_MC2_155 LQALLRELVSDIGERVHLRSEAT--GRQRHDVSVISAPVHDHHQ-RQVMV
TH_0373|M.thermoresistible__bu LRALFGEMVSDIGERVYLRSEHSPAGGAKFDISVISAPVFDRDH-RQVMV
Mflv_1528|M.gilvum_PYR-GCK LRALLGELVSDIGERVYLREESS--GRRKHDISVISAPVFDHYQ-RQVMV
Mvan_5230|M.vanbaalenii_PYR-1 LRALLGELVSDIGERVYLPDEKP--GRKRHDISVISAPVFDHYG-RQVMV
MMAR_3507|M.marinum_M ITTQLARAYADLAIIDVLDEELT--SGATHSVATISAPIRGPDG-SATMT
MUL_2762|M.ulcerans_Agy99 ITTQLARAYADLAIIDVLDEELT--SGATHSVATISAPIRGPDG-SATMT
MAV_5001|M.avium_104 PTAQVARRLTTLMTMVTLRSVTDVLETELPTTEFVVLPIFGHDG-QPEYQ
MAB_3479c|M.abscessus_ATCC_199 RLDTIQSMLATLLARMGEPIDFS----EPRRIGFLGAPVFDGNGEVSVML
. : : : *: .
MSMEG_5933|M.smegmatis_MC2_155 VSLHVGRALTDSEITKRARGVVAAADAVTAQLGGVKPTR-----------
TH_0373|M.thermoresistible__bu TSLHVGDALSENEIRRRARAVVATADAVTRQLGGAKPRWAGLTDPGRRVR
Mflv_1528|M.gilvum_PYR-GCK TSMHIGKALTDNEISERARAVVAAADAVTAQLGGMRP--------RDPD-
Mvan_5230|M.vanbaalenii_PYR-1 ASMHIGKALTDNEIRERAKALIGTADAVTAQLGGARPTSWPHTVGKGPDR
MMAR_3507|M.marinum_M VMATVFANLDGAGIRKLGERLRRSAHAIEQRSARSSA-------------
MUL_2762|M.ulcerans_Agy99 VMATVFANLDGAGIRKLGERLRRSAHAIEQRSARSSA-------------
MAV_5001|M.avium_104 IEIHLGHSAG-LTLPELDDALEQARRLLTAPVR-----------------
MAB_3479c|M.abscessus_ATCC_199 SVLGTPNRLTEAEVAHAGNQLRFCADHITSITHGRQGSGT----------
: . : :
MSMEG_5933|M.smegmatis_MC2_155 --
TH_0373|M.thermoresistible__bu --
Mflv_1528|M.gilvum_PYR-GCK --
Mvan_5230|M.vanbaalenii_PYR-1 DH
MMAR_3507|M.marinum_M --
MUL_2762|M.ulcerans_Agy99 --
MAV_5001|M.avium_104 --
MAB_3479c|M.abscessus_ATCC_199 --