For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQTIALIGFLGGLITGISPCILPVLPVILLSGMDGDRRGVGSAARPYLVIAGLVCSFSVATLIGSALLTA LHLPQDAIRWTALVVLTAIGLGLIFPPLQQLIERPFAYLPQRQSRPGADGFGLGLTLGALYVPCAGPVLA AIVVAGGTSSIGPGALVLTATFAAGNALPLLAFALAGRRVAQRVAAFRRRQRAIQVAGGIAMIVLAVALV FNLPAMLQRAVPDYTTAMQNRLGANDLQRSLSPPGPPPTQGGALQLSPGDVTGGGALSDCSDGLAELQRC GPAPALTGITGWLNTPDGKPLDPAAVRGKVILIDFWAYSCINCQRAIPHVIDWYDRYHDSGFLVIGVHTP EYAFERVPGNVASGAADLHIGYPIALDNDYATWNNYQNLYWPAEYLIDATGQVRHTKFGEGDYDGTERLI RELLAAAHPGARLPAPANTADTTPQSRLTPETYLGVGKAGNYGGTGDYRSGTATLSYPATLGEDRFALRG RWTLDDQGATAAGDDCAVRLNYTAKDVYAVVGGTGTLTVTRDGTTTTTPIGGAPTLHRIVADDSAHRDQL DMRVSPGLQVFSFTFG
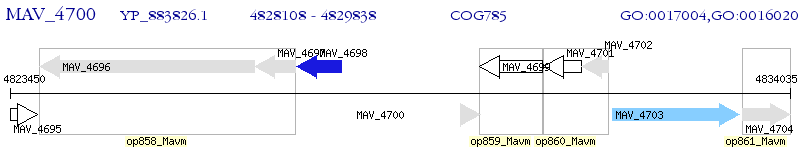
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4700 | - | - | 100% (576) | DipZ protein |
| M. avium 104 | MAV_4618 | - | 3e-13 | 30.09% (226) | cytochrome C biogenesis protein transmembrane region |
| M. avium 104 | MAV_3728 | - | 1e-05 | 26.29% (213) | cytochrome C biogenesis protein transmembrane region |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2899 | dipZ | 0.0 | 61.74% (596) | integral membrane C-type cytochrome biogenesis protein DipZ |
| M. gilvum PYR-GCK | Mflv_2825 | - | 6e-16 | 27.15% (221) | cytochrome c biogenesis protein, transmembrane region |
| M. tuberculosis H37Rv | Rv2874 | dipZ | 0.0 | 61.58% (596) | integral membrane C-type cytochrome biogenesis protein DipZ |
| M. leprae Br4923 | MLBr_02411 | - | 5e-12 | 25.68% (222) | putative cytochrome C-type biogenesis protein |
| M. abscessus ATCC 19977 | MAB_3975c | - | 4e-17 | 30.88% (217) | putative cytochrome C biogenesis protein CcdA |
| M. marinum M | MMAR_1835 | dipZ | 0.0 | 61.29% (589) | integral membrane C-type cytochrome biogenesis protein DipZ |
| M. smegmatis MC2 155 | MSMEG_0972 | - | 2e-14 | 28.32% (226) | cytochrome C biogenesis protein transmembrane region |
| M. thermoresistible (build 8) | TH_2250 | ccsA | 4e-13 | 27.92% (240) | POSSIBLE CYTOCHROME C-TYPE BIOGENESIS PROTEIN CCDA |
| M. ulcerans Agy99 | MUL_2084 | dipZ | 0.0 | 60.95% (589) | integral membrane C-type cytochrome biogenesis protein DipZ |
| M. vanbaalenii PYR-1 | Mvan_0867 | - | 5e-16 | 30.67% (225) | cytochrome c biogenesis protein, transmembrane region |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3975c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 MVESRRAAAAASAYASRCGIAPATSQRSLATPPTISVPSGEGRCRCHVAR
Rv2874|M.tuberculosis_H37Rv MVESRRAAAAASAYASRCGIAPATSQRSLATPPTISVPSGEGRCRCHVAR
MMAR_1835|M.marinum_M --------------------------------------------------
MUL_2084|M.ulcerans_Agy99 --------------------------------------------------
MAV_4700|M.avium_104 --------------------------------------------------
MAB_3975c|M.abscessus_ATCC_199 ---------------------------------------------MILAD
Mvan_0867|M.vanbaalenii_PYR-1 ---------------------------------------------MTLD-
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 GAGRDPRRRLRRRRWCGRCGYHSHLTGGEFDVNRLCQQRSRERSCQLVAV
Rv2874|M.tuberculosis_H37Rv GAGRDPRRRLRRRRWCGRCGYHSHLTGGEFDVNRLCQQRSRERSCQLVAV
MMAR_1835|M.marinum_M -----------------------------------------MRPGQPAAS
MUL_2084|M.ulcerans_Agy99 --------------------------------------------------
MAV_4700|M.avium_104 --------------------------------------------------
MAB_3975c|M.abscessus_ATCC_199 AGSAFQDAATSGPLLLALGACALAGLVSFASPCVIPLVPGYLSYLAAVVG
Mvan_0867|M.vanbaalenii_PYR-1 ---QIDQLTATGPLLLATGLAALAGLVSFASPCVIPLVPGYLSYLAAVVG
MSMEG_0972|M.smegmatis_MC2_155 -MTGFAEIAAAGPVLLALGVSVLAGLVSFASPCVVPLVPGYLSYLAAVVG
TH_2250|M.thermoresistible__bu -MTGFAELAAAGPVLVAVAISILAGLVSFASPCVVPLVPGYLSYLAAVVG
MLBr_02411|M.leprae_Br4923 -MTGFTEIAAAGPLLVALGVCMLAGLVSFVSPCVVPLVPGYLSYLAAVVG
Mflv_2825|M.gilvum_PYR-GCK -MDGVTAIATSGHVLLAVFVSALAGLVSFASPCVVPLVPGYLSYLAAVVG
Mb2899|M.bovis_AF2122/97 PADPRPKRQRITDVLTLALVGFLGGLITGISPCILPVLP--VIFFSGAQS
Rv2874|M.tuberculosis_H37Rv PADPRPKRQRITDVLTLALVGFLGGLITGISPCILPVLP--VIFFSGAQS
MMAR_1835|M.marinum_M TSHPGPPQFRIVNMVTLALVGFVGGLITGISPCILPVLP--VVFFSGAH-
MUL_2084|M.ulcerans_Agy99 -----------MNMVTLALVGFVGGLITGISPCILPVLP--VVFFSGAH-
MAV_4700|M.avium_104 -------------MQTIALIGFLGGLITGISPCILPVLP--VILLSGMD-
: :.**:: ***::*::* : ::.
MAB_3975c|M.abscessus_ATCC_199 ADNGPENGAG----------------AQPHRLRVTGSALLFVAGFTAVFL
Mvan_0867|M.vanbaalenii_PYR-1 VDQSAAGSAAPGAV-----------AVKGARLRVAGAAGLFVAGFTVVFL
MSMEG_0972|M.smegmatis_MC2_155 VDDD--PVRTGGER----------VALRSARLRVAGAAALFVAGFTVVFL
TH_2250|M.thermoresistible__bu VPKEAAPVVTGGPRRDPPADSGAGMSPRSARLRVTGAAALFVAGFTVVFL
MLBr_02411|M.leprae_Br4923 VSHEETQPGAGVIK-----------TPPAARWRVAGSAVLFVAGFTTVFV
Mflv_2825|M.gilvum_PYR-GCK VDDDQARAQP----------------LGRARLRVAGAAGLFVVGFTAVFL
Mb2899|M.bovis_AF2122/97 VDAAQVAKPEGAVA-----------VRRKRALSATLRPYRVIGGLVLSFG
Rv2874|M.tuberculosis_H37Rv VDAAQVAKPEGAVA-----------VRRKRALSATLRPYRVIGGLVLSFG
MMAR_1835|M.marinum_M -GGATVATTQTGVS-----------TPDPKAARVQTRPYRVIAGLVLSFC
MUL_2084|M.ulcerans_Agy99 -GGATVATTQTGVS-----------TPDPKAARVQTRPYRVIAGLVLSFC
MAV_4700|M.avium_104 --------------------------GDRRGVGSAARPYLVIAGLVCSFS
. .: *:. *
MAB_3975c|M.abscessus_ATCC_199 LGTVAVMGMTTSLIT--NQVLLQRIGGVVTIAMGLVFIGLVPALQRDIRF
Mvan_0867|M.vanbaalenii_PYR-1 LGTVAVLGMTTALIT--NQLLLQRLGGVVTILMGLVFIGLVPVLQRDTRF
MSMEG_0972|M.smegmatis_MC2_155 LGAVAVLGMTTTLIA--NQLLLQRIGGVVTIVMGLVFVGFIPALQRQARF
TH_2250|M.thermoresistible__bu LGTVAVLGMTTTLIT--NQVLLQRIGGVITIVMGLVFIGFIPALQRQARF
MLBr_02411|M.leprae_Br4923 LDTVAVLGMTTVLTT--HQVLLQRVGGVLTIVMGLVFVGLLPALQRQVQF
Mflv_2825|M.gilvum_PYR-GCK MGTVAILGLTTTIIT--NGVLLQRIGGVVTIVMGLVFMGFFPALQRDARF
Mb2899|M.bovis_AF2122/97 MVTLLGSALLSVLHLPQDAIRWAALVALVAIGAGLIFPRFEQLLEKPFSR
Rv2874|M.tuberculosis_H37Rv MVTLLGSALLSVLHLPQDAIRWAALVALVAIGAGLIFPRFEQLLEKPFSR
MMAR_1835|M.marinum_M LVTLTGSALLSLLHLRQDTIRWVALAALVAIGLGLIFPRLEQWLQRPFSR
MUL_2084|M.ulcerans_Agy99 LVTLTGSALLSLLHLRQDTIRWVALAALVAIGLGLIFPRLEQWLQRPFSR
MAV_4700|M.avium_104 VATLIGSALLTALHLPQDAIRWTALVVLTAIGLGLIFPPLQQLIERPFAY
: :: .: : : . : : : :* **:* : :::
MAB_3975c|M.abscessus_ATCC_199 TPRRVSTVAGAPL-LGAVFALGWTPCLGPTLTGVVAVASATEGADVARGI
Mvan_0867|M.vanbaalenii_PYR-1 TPRQISTLGGAPL-LGAVFALGWTPCLGPTLTGVIAVASATEGSNVARGV
MSMEG_0972|M.smegmatis_MC2_155 TPRQLSTVAGAPL-LGAVFALGWTPCLGPTLTGVIAVASATDGATVARGV
TH_2250|M.thermoresistible__bu APRQWSTVAGAPL-LGAVFALGWTPCLGPTLTGVITVASATDGASVARGV
MLBr_02411|M.leprae_Br4923 SLRQLTTVAGAPV-LGTVFALGWTPCLGPTLSGVITVAAATDGVNVTRGI
Mflv_2825|M.gilvum_PYR-GCK TPTSLSTLWGAPL-LGAVFGLGWTPCLGPTLTGVIAVASATEGPNVVRGI
Mb2899|M.bovis_AF2122/97 IPQKQIVTRSNGFGLGLALGVLYVPCAGPILAAIVVAGATATIG--LGTV
Rv2874|M.tuberculosis_H37Rv IPQKQIVTRSNGFGLGLALGVLYVPCAGPILAAIVVAGATATIG--LGTV
MMAR_1835|M.marinum_M IPHNSISAGGTGFGLGLALGVLYVPCAGPVLTAIILAGATSRIG--LNTV
MUL_2084|M.ulcerans_Agy99 IPHNSISAGGTGFGLGLALGVLYVPCAGPVLTAIILAGATSRIG--LNTV
MAV_4700|M.avium_104 LPQRQSRPGADGFGLGLTLGALYVPCAGPVLAAIVVAGGTSSIG--PGAL
. . ** .:. :.** ** *:.:: ...:: :
MAB_3975c|M.abscessus_ATCC_199 ALVIAYCLGLGMPFLLLAFGSARAVRGMDWLRRHTRMIQVIGGIALLAVG
Mvan_0867|M.vanbaalenii_PYR-1 VLVIAYCLGLGIPFVLLAFGSARAVAGLGWLRRHTRTIQVLGGVLMILVG
MSMEG_0972|M.smegmatis_MC2_155 VLVLAYCLGLGIPFVLLALGSARAVQGLGWLRRHTRTIQIFGGVLLILVG
TH_2250|M.thermoresistible__bu VLVIAYCLGLGIPFILLAFGSARAVQGLGWLRRNTRTIQIFGGVLLILVG
MLBr_02411|M.leprae_Br4923 LLVIAYCMGLGIPFVLLASGSAQAVAGLRWLRQYGRAIQVFGGMLLIAIG
Mflv_2825|M.gilvum_PYR-GCK VLVVAYCVGLGAPFVLFALASGRAMRALGWLRRNARRIQVFGGVLLVAVG
Mb2899|M.bovis_AF2122/97 VLTATFALGAALPLLFFALAGQRIAERVGAFRRRQREIRIATGSVTILLA
Rv2874|M.tuberculosis_H37Rv VLTATFALGAALPLLFFALAGQRIAERVGAFRRRQREIRIATGSVTILLA
MMAR_1835|M.marinum_M VLTCAFALGTALPLLFFALAGRETAGRVSAFRRRQREIRIGAGVVTILLA
MUL_2084|M.ulcerans_Agy99 VLTCAFALGTALPLLFFALAGRETAGRVSAFRRRQHEIRIGAGVVTILLA
MAV_4700|M.avium_104 VLTATFAAGNALPLLAFALAGRRVAQRVAAFRRRQRAIQVAGGIAMIVLA
*. ::. * . *:: :* .. . : :*: : *:: * : :.
MAB_3975c|M.abscessus_ATCC_199 VALITGVWADFVAWVRDSFVSDARLPI-----------------------
Mvan_0867|M.vanbaalenii_PYR-1 AALVTGLWNDFVSWIRDAFVSDVRLPI-----------------------
MSMEG_0972|M.smegmatis_MC2_155 AALVTGLWNEFVSYVRDAFVSDVRLPI-----------------------
TH_2250|M.thermoresistible__bu AALVTGLWNEFISWVRDAFVSDVRLPI-----------------------
MLBr_02411|M.leprae_Br4923 AALIAGVWDDFVSWLRDAVVSDMRVSI-----------------------
Mflv_2825|M.gilvum_PYR-GCK IALLTGLWNEVVSWVRDAFVSNTTLPI-----------------------
Mb2899|M.bovis_AF2122/97 VALVFDLPAALQRAIPDYTASLQ------------QQISTGTEIREQLN-
Rv2874|M.tuberculosis_H37Rv VALVFDLPAALQRAIPDYTASLQ------------QQISTGTEIREQLN-
MMAR_1835|M.marinum_M VALVLNLPATLQRAIPDYTSALQGLP---------EQVGGPNQIGTNLGG
MUL_2084|M.ulcerans_Agy99 VALVLNLPATLQRAIPDYTSALQGLP---------EQVGGPNQIGTNLGG
MAV_4700|M.avium_104 VALVFNLPAMLQRAVPDYTTAMQNRLGANDLQRSLSPPGPPPTQGGALQL
**: .: . : * :
MAB_3975c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 LGGIVNAQNAQLSNCSDGAAQLESCGTAPDLKGITGWLNTPGNKPIDLKS
Rv2874|M.tuberculosis_H37Rv LGGIVNAQNAQLSNCSDGAAQLESCGTAPDLKGITGWLNTPGNKPIDLKS
MMAR_1835|M.marinum_M IGGLVTDQNAQLSNCTNGAAQPQDCGSAPDITGISGWLNTPEHQPIRLSS
MUL_2084|M.ulcerans_Agy99 IGGLVTDQNAQLSNCTNGAAQPQDCGSAPDITGISGWLNTPEHQPIRLSS
MAV_4700|M.avium_104 SPGDVTGG-GALSDCSDGLAELQRCGPAPALTGITGWLNTPDGKPLDPAA
MAB_3975c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 LRGKVVLIDFWAYSCINCQRAIPHVVGWYQAYKDSGLAVIGVHTPEYAFE
Rv2874|M.tuberculosis_H37Rv LRGKVVLIDFWAYSCINCQRAIPHVVGWYQAYKDSGLAVIGVHTPEYAFE
MMAR_1835|M.marinum_M LRGRVVLVDFWAYSCINCQRAIPHVVGWYRAYKDSGLEVIGVHTPEYAFE
MUL_2084|M.ulcerans_Agy99 LRGRVVLVDFWAYSCINCQRAIPHVVGWYRAYKDSGLEVIGVHTPEYAFE
MAV_4700|M.avium_104 VRGKVILIDFWAYSCINCQRAIPHVIDWYDRYHDSGFLVIGVHTPEYAFE
MAB_3975c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 KVPGNVAKGAANLGISYPIALDNNYATWTNYRNRYWPAEYLIDATGTVRH
Rv2874|M.tuberculosis_H37Rv KVPGNVAKGAANLGISYPIALDNNYATWTNYRNRYWPAEYLIDATGTVRH
MMAR_1835|M.marinum_M KVPENVAAGAADLNITYPIALDNNYSTWTNYRNAYWPAEYLIDASGTVRH
MUL_2084|M.ulcerans_Agy99 KVPENVAAGAADLNITYPIALDNNYSTWTNYRNAYWPAEYLIDASGTVRH
MAV_4700|M.avium_104 RVPGNVASGAADLHIGYPIALDNDYATWNNYQNLYWPAEYLIDATGQVRH
MAB_3975c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 IKFGEGDYNVTETLVRQLLNDAKPGVKLPQPSSTTTPDLTPRAALTPETY
Rv2874|M.tuberculosis_H37Rv IKFGEGDYNVTETLVRQLLNDAKPGVKLPQPSSTTTPDLTPRAALTPETY
MMAR_1835|M.marinum_M IKFGEGDYDGTENLIRRLLTDANPGAKLPAP--VDAADTTPALSLTPETY
MUL_2084|M.ulcerans_Agy99 IKFGEGDYDGTENLIRRLLTDANPGAKLPAP--VDAADTTPALSLTPETY
MAV_4700|M.avium_104 TKFGEGDYDGTERLIRELLAAAHPGARLPAP--ANTADTTPQSRLTPETY
MAB_3975c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 FGVGKVVNYGGGGAYDEGSAVFDYPPSLAANSFALRGRWALDYQGATSDG
Rv2874|M.tuberculosis_H37Rv FGVGKVVNYGGGGAYDEGSAVFDYPPSLAANSFALRGRWALDYQGATSDG
MMAR_1835|M.marinum_M FGVGKAVNFAGSG-YDQGSATFSYPLALPPDTFALQGPWSLDHQRATADG
MUL_2084|M.ulcerans_Agy99 FGVGKAVTFAGSG-YDQGSATFSYPLALPPDTFALQGPWSLDHQRATADG
MAV_4700|M.avium_104 LGVGKAGNYGGTGDYRSGTATLSYPATLGEDRFALRGRWTLDDQGATAAG
MAB_3975c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0972|M.smegmatis_MC2_155 --------------------------------------------------
TH_2250|M.thermoresistible__bu --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mflv_2825|M.gilvum_PYR-GCK --------------------------------------------------
Mb2899|M.bovis_AF2122/97 NDAAIKLNYHAKDVYIVVGGTGTLTVVRDGKPATLPISGPPTTHQVVAGD
Rv2874|M.tuberculosis_H37Rv NDAAIKLNYHAKDVYIVVGGTGTLTVVRDGKPATLPISGPPTTHQVVAGY
MMAR_1835|M.marinum_M DDCRIKLNYLAKNVYLVVGGTGTLTVTGAGKSTTLRIGGAPTSHQIVAGD
MUL_2084|M.ulcerans_Agy99 DDCRIKLNYLAKNVYLVVGGTGTLTVTGAGKSTTLRIGGAPTSHQIVAGD
MAV_4700|M.avium_104 DDCAVRLNYTAKDVYAVVGGTGTLTVTRDGTTTTTPIGGAPTLHRIVADD
MAB_3975c|M.abscessus_ATCC_199 -----------------------
Mvan_0867|M.vanbaalenii_PYR-1 -----------------------
MSMEG_0972|M.smegmatis_MC2_155 -----------------------
TH_2250|M.thermoresistible__bu -----------------------
MLBr_02411|M.leprae_Br4923 -----------------------
Mflv_2825|M.gilvum_PYR-GCK -----------------------
Mb2899|M.bovis_AF2122/97 RLASETLEVRPSKGLQVFSFTYG
Rv2874|M.tuberculosis_H37Rv RLASETLEVRPSKGLQVFSFTYG
MMAR_1835|M.marinum_M QVQRTTIEVHPSKGLQVYSFTYG
MUL_2084|M.ulcerans_Agy99 QVQRTTIEVHPSKGLQVYSFTYG
MAV_4700|M.avium_104 SAHRDQLDMRVSPGLQVFSFTFG