For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVVGRDLRADMRITHPLISRAHLLLRYDQGKWLAIDNGSLNGTFVNGRRVPVVEIHDGCAINIGNPDGPL LTFDVGRHQGMAGRPPRTESMGIPVAAHPPAPAAWSAQPAPPVAAPPPPAGRTPNWAAPPRRPQPAPRPG PPGQTMYGGRPPGGYPPAAPPPPPVAPAPAAPHTQMAPAASKPPEVANLATKMFQALMPSRSGAIQKPAG SQTIGRATDNDIVIQDVLASRHHAFLIQTPLGTEIRDAHSVNGTFVNGVRVGSAVLTEGDVVTIGNVDLV FTRDGLIRRTEAATRSGGLEVNSVCYTVDHGKQLLDHISLTARPGTLTAIIGGSGAGKTTLSRLIVGYTT PTSGTVTFEGHNIHTEYASMRSRIGMVPQDDVVHRQLTVNQALGYAAELRLPPDTSKEERARVVAQVLEE LDMTQHAETRVDKLSGGQRKRASVALELLTQPSLLLLDEPTSGLDPALDRQVMLMLRQLADAGRVVLVVT HSVSYLDVCDQILLIAPGGKTAFCGPPDQVEPALGTRNWADIFAKVGADPDEANRRFKERNQQSSQPPTP QSPADLGEPPQTDLWRQLSTIARRQVRLVVSDRGYTIFLAVLPFLIGALSLTVKGPHPGLGPADPLGPAP TQPQYIMVLLNIGAVFMGTALTIRDLIGERAIFRREQAVGLSTTAYLLAKIAVFCVFATAQAAIATIIVR LGKGAPTAHPQFFGNSTFSLFVTVAGTCIASAMLGLLLSALAQSNEQIMPLLVVSIMSQLVFSSGMIPVY QRIGLEQLAWLTPARWGYAAGASSIDFPSLVKVKQIPTNDPIWQHSKHIFVFDMAMLALLSIGYTGFVRW HIRLKR
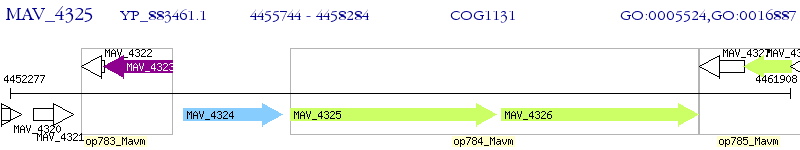
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4325 | - | - | 100% (846) | ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_3010 | - | 0.0 | 63.56% (859) | ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_3146 | - | 0.0 | 63.01% (857) | ABC transporter, ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1776 | - | 0.0 | 62.82% (858) | transmembrane ATP-binding protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_4882 | - | 0.0 | 62.17% (859) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1747 | - | 0.0 | 62.82% (858) | transmembrane ATP-binding protein ABC transporter |
| M. leprae Br4923 | MLBr_00669 | ftsE | 4e-22 | 31.53% (203) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_3698 | - | 0.0 | 54.44% (856) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_1174 | - | 0.0 | 72.38% (858) | transmembrane ATP-binding protein ABC transporter |
| M. smegmatis MC2 155 | MSMEG_1642 | - | 0.0 | 61.61% (883) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4260 | - | 0.0 | 67.90% (673) | PUTATIVE ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_3149 | - | 0.0 | 62.20% (865) | transmembrane ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_1546 | - | 0.0 | 62.38% (864) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4325|M.avium_104 ---------------------------------MVVGRDLRADMRITHPL
MMAR_1174|M.marinum_M --------MPQPAPPALTVRCDGSERTFAAGHDVVVGRDLRADMRITHPL
MSMEG_1642|M.smegmatis_MC2_155 -------MSRPSPP-ALTVRYEGSTRTFAPGSDVVIGRDLRADVRIAHPL
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK -------MSRPGSP-ALTVRYDGSTHTFAAGNDVVVGRDLRADVRIAHPL
Mvan_1546|M.vanbaalenii_PYR-1 -------MSRPATP-ALTVRYDGSTRSFAAGNDVVVGRDLRADIRIAHPL
Mb1776|M.bovis_AF2122/97 -----MPMSQPAAPPVLTVRYEGSERTFAAGHDVVVGRDLRADVRVAHPL
Rv1747|M.tuberculosis_H37Rv -----MPMSQPAAPPVLTVRYEGSERTFAAGHDVVVGRDLRADVRVAHPL
MUL_3149|M.ulcerans_Agy99 MPLQLVPMTQTAAPPVLTVRYNGAERTFAAGHDVVIGRDLRADVRVADPL
MAB_3698|M.abscessus_ATCC_1997 -------MTGREAP-TLTVVFDGDERKFAPGHDIHIGRDIHADVRITHPL
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 ISRAHLLLRYDQGKWLAIDNGSLNGTFVNGRRVPVVEIHDGCAINIGNPD
MMAR_1174|M.marinum_M ISRAHLLLRFDQGRWLAIDNNSLNGTFVNGRRVPMVEIYDGQSLNIGNPD
MSMEG_1642|M.smegmatis_MC2_155 ISRAHLVLRFDQGRWVAIDNGSLNGMYVNGRRVSSVDLQDGQVLNIGNPD
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK ISRAHLVLRFDQGRWIAIDNGSLNGMYVNGRRVPAADISDGGQLHIGNPD
Mvan_1546|M.vanbaalenii_PYR-1 ISRAHLVLRFDQGRWLAIDNGSLNGMYVNGRRVPAAEITDGTQVHIGNPD
Mb1776|M.bovis_AF2122/97 ISRAHLLLRFDQGRWVAIDNGSLNGLYLNNRRVPVVDIYDAQRVHIGNPD
Rv1747|M.tuberculosis_H37Rv ISRAHLLLRFDQGRWVAIDNGSLNGLYLNNRRVPVVDIYDAQRVHIGNPD
MUL_3149|M.ulcerans_Agy99 ISRVHLLLRFDQGRWIAIDNGSLNGLYINNRRVPVVDIQDGQRVNIGNPD
MAB_3698|M.abscessus_ATCC_1997 VSRIHLIIRFVDGKWIAEDQRSLNGIFVNGRQVGWVEVRDDTVINLGDPG
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 GPLLTFDVG-RHQGMAGRPPRTESMGIPVAAHPPAPAAWSAQPAPPVAAP
MMAR_1174|M.marinum_M GPLLSFEVG-RHVGMAGRPPQTESMRVTPSMVRPS------GQHPVAHGP
MSMEG_1642|M.smegmatis_MC2_155 GPQLSFEVG-RHQGSAGRTP-TAAVPVAG-HTGTS----WPTQAPTGGGW
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK GPLLTFEVG-RHQGSAGTPPSTTAVPVPG-RTSAT----WPAQHS-GPTG
Mvan_1546|M.vanbaalenii_PYR-1 GPLLVFEVG-RHQGSAGSPPPTAAVPVAG-RPSAT----WPSQP------
Mb1776|M.bovis_AF2122/97 GPALDFEVG-RHRGSAGRPPQTTSIRLPN-LSAGA----WPTDGP--PQT
Rv1747|M.tuberculosis_H37Rv GPALDFEVG-RHRGSAGRPPQTTSIRLPN-LSAGA----WPTDGP--PQT
MUL_3149|M.ulcerans_Agy99 GPALDFGVGRRQQGSVGRPPPTAVMQVPPPMPSGP----WPSQAP--PQT
MAB_3698|M.abscessus_ATCC_1997 GPRLTFALG----GLGGETQLGIAPPTMRGYVSGA------QQQAPYTGA
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 PPPAGRTPNWAAPPRRPQP-APRPGPP--GQTMYGGR----PPGGYPPAA
MMAR_1174|M.marinum_M TSTGSHTSHEAPPTARFHPGAAPPGPARTGQPMYPSSSAPWRPTGYPASP
MSMEG_1642|M.smegmatis_MC2_155 QQPYTQPPRTQYPQTTGTQQRYPSAPQHGYPNGPQTGYPSGPQRGYPSGP
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK RPPVQRPPTGPYPGAGAYPPGGPRPVPAPPPPAAQ-SRPAHVQQPISAPA
Mvan_1546|M.vanbaalenii_PYR-1 -PPSQPPPTGRPPAQRPYP-SAPRPSAPPTYPTPQPSRPAPVQPPLSSPA
Mb1776|M.bovis_AF2122/97 GTLGSGQLQQLPPATTRIPAAPPSGP--------QPRYPTGGQQLWPPSG
Rv1747|M.tuberculosis_H37Rv GTLGSGQLQQLPPATTRIPAAPPSGP--------QPRYPTGGQQLWPPSG
MUL_3149|M.ulcerans_Agy99 GSHRPVPPQPAPP-TTRMPAQPPSGPQHVQPSDQQPQYPSGPQTSRSQTG
MAB_3698|M.abscessus_ATCC_1997 QQHGTTGQPYHSTQQHYPTGGYQSGPQTYGQPPSQQHPYGAAPSGPQTYG
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 PPPPP--VAPAPAAPHTQMAPAASKPPE----------------------
MMAR_1174|M.marinum_M APPPPSNFPPPPPRPGPQPPQQATKPPE----------------------
MSMEG_1642|M.smegmatis_MC2_155 QTGYPTNG--PAGAPQSYQSQPVRTPPPAPANSSQAPTTMGPAAAPRGGA
TH_4260|M.thermoresistible__bu ----------------------VRSPE-----------------------
Mflv_4882|M.gilvum_PYR-GCK MDPVTVMG--PAAAP--------RSSGS----------------------
Mvan_1546|M.vanbaalenii_PYR-1 MESATVMG--PAAAP--------RSGG-----------------------
Mb1776|M.bovis_AF2122/97 PQRAPQIYRPPTAAPPPAG----ARGGT----------------------
Rv1747|M.tuberculosis_H37Rv PQRAPQIYRPPTAAPPPAG----ARGGT----------------------
MUL_3149|M.ulcerans_Agy99 AQPAPQIYRPPAASPAPPAPLVVGRAPG----------------------
MAB_3698|M.abscessus_ATCC_1997 TSGPTRPRQQYPSEPTPVP---ASYSNE----------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 --VANLATKMFQALMPSRSGAIQKPAGSQTIGRATDNDIVIQDVLASRHH
MMAR_1174|M.marinum_M --VANLATKMFHALLP-RSGGAERPAGSVTIGRATDNDIVIPDVLASRHH
MSMEG_1642|M.smegmatis_MC2_155 EPASNLATSMLKILRPGRSA--PAPAGAVKIGRATDNDIVIPDVLASRHH
TH_4260|M.thermoresistible__bu --INNLATSMMKILRPGRAQ--EAPPGSIKIGRAVDNDIVIPDVLASRHH
Mflv_4882|M.gilvum_PYR-GCK --DGNIATSMLRILRPGRPA--DVPAGSIKIGRSSDNDIVIPDVLASRHH
Mvan_1546|M.vanbaalenii_PYR-1 --DGNIATSMLKILRPGRPA--EAPPGSIKIGRSTDNDIVIPDVLASRHH
Mb1776|M.bovis_AF2122/97 -EAGNLATSMMKILRPGRLTG-ELPPGAVRIGRANDNDIVIPEVLASRHH
Rv1747|M.tuberculosis_H37Rv -EAGNLATSMMKILRPGRLTG-ELPPGAVRIGRANDNDIVIPEVLASRHH
MUL_3149|M.ulcerans_Agy99 -ETGNIATSMMKILRPGRAAG-ELPPGAVTIGRADDNDIVIPEVLASRHH
MAB_3698|M.abscessus_ATCC_1997 -PVEPFGTRAMRALGIGGAP--KPTPGAITVGRAPDNTIVVNDVLASRHH
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 AFLIQTPLGTEIRDAHSVNGTFVNGVRVGSAVLTEGDVVTIGNVDLVFTR
MMAR_1174|M.marinum_M AFLTLTPLGTEIRDAHSVNGTFVNGVRVGSAVLSDGDVVTIGNVDLVFTG
MSMEG_1642|M.smegmatis_MC2_155 ATLIPLPGGTEIRDERSINGTFVNGTRVDSAVLHDGDVVTIGNVDLVFSG
TH_4260|M.thermoresistible__bu ATLVPTPAGMEIRDNRSINGTFVNGTRTETAVLNEGDVVTIGNVDLVFRN
Mflv_4882|M.gilvum_PYR-GCK ATLVPTPAGTEIRDNRSINGTFVNGSRVDSAILADGDTVTIGNVDLSFRD
Mvan_1546|M.vanbaalenii_PYR-1 ATLVHSPAGTEIRDNRSINGTFVNGARVDSAMLTEGDTVTIGNVDLVFRD
Mb1776|M.bovis_AF2122/97 ATLVPTPGGTEIRDNRSINGTFVNGARVDAALLHDGDVVTIGNIDLVFAD
Rv1747|M.tuberculosis_H37Rv ATLVPTPGGTEIRDNRSINGTFVNGARVDAALLHDGDVVTIGNIDLVFAD
MUL_3149|M.ulcerans_Agy99 ATLIPTPQGTEIRDNRSINGTFVNGARVEAALLHDGDVVTIGNIDLIFAN
MAB_3698|M.abscessus_ATCC_1997 ALLLPTPGGLEIQDLHTINGTFVNGVQVEQSSLREGDTVTIGNVDLVVQD
MLBr_00669|M.leprae_Br4923 -------------------------------------MITLDHVSKKYKS
:*:.::.
MAV_4325|M.avium_104 DGLIRR--TEAATRSGGLEVNSVCYTVDHGKQLLDHISLTARPGTLTAII
MMAR_1174|M.marinum_M GTLIRR--TEAATRSGGLEVKSVCFTVDGGKQLLDHISLTARPGTLTAII
MSMEG_1642|M.smegmatis_MC2_155 GTLARRSETEADTRTGGLEVRGLTWTIEGNKTLLDNISIDARPGTLTAVI
TH_4260|M.thermoresistible__bu GTLVRRTETEAATDAGGLDVRGVTWTIENNKTLLDNISITARPGTLTAII
Mflv_4882|M.gilvum_PYR-GCK GTLVRRTETAAATATGGLDVHGVTWTIENNKTLLDNISMSARPGTLTAVI
Mvan_1546|M.vanbaalenii_PYR-1 GTLVRRTETAAATATGGLDVHGVTWTIENNKTLLDNISMSARPGTLTAVI
Mb1776|M.bovis_AF2122/97 GTLARREENLLETRVGGLDVRGVTWTIDGDKTLLDGISLTARPGMLTAVI
Rv1747|M.tuberculosis_H37Rv GTLARREENLLETRVGGLDVRGVTWTIDGDKTLLDGISLTARPGMLTAVI
MUL_3149|M.ulcerans_Agy99 GALARREESLLETRTGGLDVHGVTWTIEGNKTLLDNISLAARPGMLTAVI
MAB_3698|M.abscessus_ATCC_1997 GTLVKP---TTATRVGGLEVQSIDFVVEGGKKLLDDISFSAGPGSLTAVI
MLBr_00669|M.leprae_Br4923 ----------------------------LARPALDNVNVKIDKGEFVFLI
: ** :.. * :. :*
MAV_4325|M.avium_104 GGSGAGKTTLSRLIVGYTTPTSGTVTFEGHNIHTEY----ASMRSRIGMV
MMAR_1174|M.marinum_M GGSGAGKTTLSRLIAGYTSPSSGTVTFEGHNIHAEY----ASMRSRIGMV
MSMEG_1642|M.smegmatis_MC2_155 GPSGAGKSTFARQVAGYTHPTSGTIKFEGHDVHAEY----ASLRSRIGMV
TH_4260|M.thermoresistible__bu GPSGAGKSTFARLIAGYTHPTSGTVTFEGHNVHAEY----ASLRSRIGMV
Mflv_4882|M.gilvum_PYR-GCK GPSGAGKSTFARLVAGYTHPTTGQVSFEGHDVHAEY----ASLRSRIGMV
Mvan_1546|M.vanbaalenii_PYR-1 GPSGAGKSTFARLVAGYTHPTTGQVSFEGHDVHAEY----ASLRSRIGMV
Mb1776|M.bovis_AF2122/97 GPSGAGKSTLARLVAGYTHPTDGTVTFEGHNVHAEY----ASLRSRIGMV
Rv1747|M.tuberculosis_H37Rv GPSGAGKSTLARLVAGYTHPTDGTVTFEGHNVHAEY----ASLRSRIGMV
MUL_3149|M.ulcerans_Agy99 GPSGAGKSTFARLVAGYTHPTNGTVTFEGHNIHAEY----ASLRSRIGMV
MAB_3698|M.abscessus_ATCC_1997 GPSGAGKSTLARLIVGNTQPSAGKVSFEGHGVHAEY----AVMRNRIGMV
MLBr_00669|M.leprae_Br4923 GPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRLRQVIGCV
* **:**:*: * : * *: * : .. . :: . :*. ** *
MAV_4325|M.avium_104 PQDDVVHRQLTVNQALGYAAELRLPPDTSKEERARVVAQVLEELDMTQHA
MMAR_1174|M.marinum_M PQDDVVHRQLTVSQALGYAAELRLPPDTSKEDRAQVVAQVLEELELTKHA
MSMEG_1642|M.smegmatis_MC2_155 PQDDVVHGQLTVRQALMYAAELRLPPDTTKEDREQVVMQVLEELEMTKHL
TH_4260|M.thermoresistible__bu PQDDVVHGQLTVNQALMYAAELRLPPDTTEEDRRQVVAEVLEELEMTAHA
Mflv_4882|M.gilvum_PYR-GCK PQDDVVHGQLTVRQALMYAAELRLPPDTTKADREQVVNEVLEELEMTKHL
Mvan_1546|M.vanbaalenii_PYR-1 PQDDVVHGQLTVRQALMYAAELRLPPDTTKADREQVVNEVLEELEMTKHL
Mb1776|M.bovis_AF2122/97 PQDDVVHGQLTVKHALMYAAELRLPPDTTKDDRTQVVARVLEELEMSKHI
Rv1747|M.tuberculosis_H37Rv PQDDVVHGQLTVKHALMYAAELRLPPDTTKDDRTQVVARVLEELEMSKHI
MUL_3149|M.ulcerans_Agy99 PQDDVVHGQLTVRQALMYAAELRLPPDTTKEDREHVVARVLEELEMSKHL
MAB_3698|M.abscessus_ATCC_1997 PQDDVVHPQLTVSQALHYAAELRLPPDTTKEDREQVVSRVLEELEMTKHA
MLBr_00669|M.leprae_Br4923 FQDFRLLQQKTVYENVAFALEVIG---RRSDAINQVVPDVLETVGLSGKA
** : * ** . : :* *: . :** *** : :: :
MAV_4325|M.avium_104 ETRVDKLSGGQRKRASVALELLTQPSLLLLDEPTSGLDPALDRQVMLMLR
MMAR_1174|M.marinum_M ETRVDKLSGGQRKRASVALELLTGPSLLILDEPTTGLDPALDRQVMLMLR
MSMEG_1642|M.smegmatis_MC2_155 DTRVDKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLR
TH_4260|M.thermoresistible__bu DTRVDKLSGGQRKRASVAMELLTGPSLLILDEPTSGLDPALDRQVMTMLR
Mflv_4882|M.gilvum_PYR-GCK DTRVDKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLR
Mvan_1546|M.vanbaalenii_PYR-1 DTRVDKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLR
Mb1776|M.bovis_AF2122/97 DTRVDKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLR
Rv1747|M.tuberculosis_H37Rv DTRVDKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLR
MUL_3149|M.ulcerans_Agy99 ETRVDKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLR
MAB_3698|M.abscessus_ATCC_1997 ETRVDKLSGGQRKRASVAMELLTGPSLLILDEPTSGLDPALDRQVMSLMR
MLBr_00669|M.leprae_Br4923 NRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLE
: *:****:::*.::* ::. * :*: **** .*** .:::* ::.
MAV_4325|M.avium_104 QLADAGRVVLVVTHSVSYLDVCDQILLIAPGGKTAFCGPPDQVEPALGTR
MMAR_1174|M.marinum_M QLADAGRVVLIVTHSVAYLDVCDQLLLVAPGGKTAFLGPPSEISQAMGTA
MSMEG_1642|M.smegmatis_MC2_155 QLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFFGPPSQIGPELGTT
TH_4260|M.thermoresistible__bu QLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGPTMGTT
Mflv_4882|M.gilvum_PYR-GCK QLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGSAMGTT
Mvan_1546|M.vanbaalenii_PYR-1 QLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGPSMGTT
Mb1776|M.bovis_AF2122/97 QLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPTQIGPVMGTT
Rv1747|M.tuberculosis_H37Rv QLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPTQIGPVMGTT
MUL_3149|M.ulcerans_Agy99 QLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPSQIGPAMGTT
MAB_3698|M.abscessus_ATCC_1997 QLADAGRVVVVVTHSLTYLSYCDQVLLLAPGGKTAYCGPPRSIGQEMGTT
MLBr_00669|M.leprae_Br4923 RINRTGTTVLMATHDHHIVDSMRQRVVELSLG-------------RLVRD
:: :* .*::.**. :. * :: . * :
MAV_4325|M.avium_104 NWADIFAKVGADPDEANRRFKERNQQSSQPPTPQSP-ADLG-EPPQTDLW
MMAR_1174|M.marinum_M NWADIFTNVGADPDEANRRFLEQKQEQPAFPSEASPAVDLG-EPVHTDVL
MSMEG_1642|M.smegmatis_MC2_155 NWADIFSTVADDPAEANRRYLARTPEAPPASASSEAPADLG-APAKTSLR
TH_4260|M.thermoresistible__bu NWADIFSTVAGDPDAANRRFLERQGPPRELPAESAKPTDLG-SPTRTSLR
Mflv_4882|M.gilvum_PYR-GCK NWADIFSSVAGDPEAAAARYLSQNGPPPPQPP-ALTPSELG-NPTRTSLR
Mvan_1546|M.vanbaalenii_PYR-1 NWADIFSAVAGDPDAAAQRYLAQNGPPPPQPP-ALTPSELG-NPTRTSLR
Mb1776|M.bovis_AF2122/97 NWADIFSTVADDPDAAKARYLARTGPTPPPPP-VEQPAELG-DPAHTSLF
Rv1747|M.tuberculosis_H37Rv NWADIFSTVADDPDAAKARYLARTGPTPPPPP-VEQPAELG-DPAHTSLF
MUL_3149|M.ulcerans_Agy99 NWADIFSTVADDPEGAQARYLAQTGPPPPAPP-VERPGELGGDPSHTSLF
MAB_3698|M.abscessus_ATCC_1997 DWADIFANAAADPDGVHQAYLSRHPAANKPATAASPPGDLG-KPPKTSLL
MLBr_00669|M.leprae_Br4923 EWCGIYGMDR----------------------------------------
:*..*:
MAV_4325|M.avium_104 RQLSTIARRQVRLVVSDRGYTIFLAVLPFLIGALSLTVKGPHPGLGPAD-
MMAR_1174|M.marinum_M HQFSTVARRQIRLVISDRGYTVFLALLPFLIGTLTLTVRG-KTGYGMSD-
MSMEG_1642|M.smegmatis_MC2_155 RQFSTIGRRQLRLIISDRGYFIFLALLPFIMGTLSLSVPG-TVGFGVPN-
TH_4260|M.thermoresistible__bu RQFSTIARRQIRLIVSDRGYFVFLALLPFIMGVLSLSVPG-DVGFGVPN-
Mflv_4882|M.gilvum_PYR-GCK RQLSTIARRQIRLVISDRGYFAFLMLLPFIMGILSLSVPG-DVGFGVPKT
Mvan_1546|M.vanbaalenii_PYR-1 RQLSTIARRQIRLVISDRGYFAFLMLLPFIMGVLSLSVPG-DVGFGVPKT
Mb1776|M.bovis_AF2122/97 RQFSTIARRQLRLIVSDRGYFVFLALLPFIMGALSMSVPG-DVGFGFPN-
Rv1747|M.tuberculosis_H37Rv RQFSTIARRQLRLIVSDRGYFVFLALLPFIMGALSMSVPG-DVGFGFPN-
MUL_3149|M.ulcerans_Agy99 RQFSTIARRQMRLIISDRGYFIFLAILPFVMGSLSMSVPG-DVGFGMPN-
MAB_3698|M.abscessus_ATCC_1997 RQVSTVARRQGRLILADRGYFIFLALLPFVLAGLVLSVPG-SAGFNVALP
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 --PLGPAPTQPQYIMVLLNIGAVFMGTALTIRDLIGERAIFRREQAVGLS
MMAR_1174|M.marinum_M --PMGNNPAQPDQILVMLNVGAVFMGTALTIRDLIGERPIFRREQAVGLS
MSMEG_1642|M.smegmatis_MC2_155 --PMGDAPNEPGQILVLLNVGAIFMGTALTIRDLIGERAIFRREQAVGLS
TH_4260|M.thermoresistible__bu --PMGDAPNEPGQILVLLNVGAIFMGTALTIRDLIGERAIFRREQAVGLS
Mflv_4882|M.gilvum_PYR-GCK ALEGGEAPNEPGQILVMLNVGAIFMGTALTIRALIGERAIFRREQAVGLS
Mvan_1546|M.vanbaalenii_PYR-1 ALEGGEAPNEPGQILVMLNVGAIFMGTALTIRALISERAIFRREQAVGLS
Mb1776|M.bovis_AF2122/97 --PMGDAPNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLS
Rv1747|M.tuberculosis_H37Rv --PMGDAPNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLS
MUL_3149|M.ulcerans_Agy99 --PMGNAPNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLS
MAB_3698|M.abscessus_ATCC_1997 -----DNPNEPGLLLALTNLGASFLGLALTIRDLIGERVIFRREQAVGLS
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 TTAYLLAKIAVFCVFATAQAAIATIIVRLGKGAP-----TAHPQFFGNST
MMAR_1174|M.marinum_M TAAYMGAKIVVFCAFAVTQAAIATTISVVGWGKP-----LSDAVLLGDVR
MSMEG_1642|M.smegmatis_MC2_155 TTAYLLAKVCVYSVFAVVQSAIVTVITISGKGWG--EGAVERGVVIPNRS
TH_4260|M.thermoresistible__bu TTAYLLAKVCVYSVFAIVQSAIVTAITIAGKGWDVDKGAVDHGSVFGNPK
Mflv_4882|M.gilvum_PYR-GCK TSAYLLAKVVVFTAFAIVQAAIVTTIAIIGKGWG--PGAVDSGAVVGDRS
Mvan_1546|M.vanbaalenii_PYR-1 TSAYLLAKVVVFTAFAIVQAAIVTSIAIAGKGWG--PGAVERGAVIGNRS
Mb1776|M.bovis_AF2122/97 TTAYLIAKVCVYTVLAVVQSAIVTVIVLVGKGGP-----TQGAVALSKPD
Rv1747|M.tuberculosis_H37Rv TTAYLIAKVCVYTVLAVVQSAIVTVIVLVGKGGP-----TQGAVALSKPD
MUL_3149|M.ulcerans_Agy99 TTAYLLAKICVYTVFAVVQSAIVTIIVLIGKGGP-----TQGAVALGKPG
MAB_3698|M.abscessus_ATCC_1997 ATAYLLAKIITYCGAAIVQSAIMTAVIVIVRGGP-----TQGAVLLGSAN
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 FSLFVTVAGTCIASAMLGLLLSALAQSNEQIMPLLVVSIMSQLVFSSGMI
MMAR_1174|M.marinum_M FELFVTVAATCVASALLGMALSSLAQSQDQIMPMLVVSIMSQLVFSGGMI
MSMEG_1642|M.smegmatis_MC2_155 LELFLSMAATTVTAAMVGLALSALAKTSEQIMPLLVVAIMSQLVFSGGMI
TH_4260|M.thermoresistible__bu LELFFGMAATTVAAAMLGLLLSALAKTSEQIMPLLVVAIMSQLVFSGGMI
Mflv_4882|M.gilvum_PYR-GCK IELFLDIAVTCVASAMVGLALSALAKSAEQIMPLLVVAIMSQLVFSGGMI
Mvan_1546|M.vanbaalenii_PYR-1 VELFTDIAVTCVAAAMVGLALSALAKSAEQIMPLLVVAIMSQLVFSGGMI
Mb1776|M.bovis_AF2122/97 LELFVDVAVTCVASAMLGLALSAIAKSNEQIMPLLVVAVMSQLVFSGGMI
Rv1747|M.tuberculosis_H37Rv LELFVDVAVTCVASAMLGLALSAIAKSNEQIMPLLVVAVMSQLVFSGGMI
MUL_3149|M.ulcerans_Agy99 LELFADVAATCVASAMLGLALSAVAKSNEQIMPLLVVAVMSQLVFSGGMI
MAB_3698|M.abscessus_ATCC_1997 FELWVAVAATACAATVLGLVLSSLATSAEQVMPMLVIATMTQIVFSGGLI
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 PVYQRIGLEQLAWLTPARWGYAAGASSIDFPSLVKVKQIPTNDPIWQHSK
MMAR_1174|M.marinum_M WVTNRFLLDQLSWLTPARWGFAASASTIDTHRLVPGPTDPK-DQHWDHKA
MSMEG_1642|M.smegmatis_MC2_155 PVTGRIGLDQLSWITPARWGFAASASTVDLIRLVPGPLTPK-DSHWEHTS
TH_4260|M.thermoresistible__bu PVTDRIGLDQLSWFTPARWGFAASASTADLITLVPGPLTPK-DRHWEHDS
Mflv_4882|M.gilvum_PYR-GCK PVTDRLVLDQMSWFTPARWGFAASASTVDLIRLVPGPLTPQ-DRHWEHTP
Mvan_1546|M.vanbaalenii_PYR-1 PVTDRIVLDQMSWFTPARWGFAASASTVDLIRLVPGPLTPQ-DRHWQHTT
Mb1776|M.bovis_AF2122/97 PVTGRVPLDQMSWVTPARWGFAASAATVDLIKLVPGPLTPK-DSHWHHTA
Rv1747|M.tuberculosis_H37Rv PVTGRVPLDQMSWVTPARWGFAASAATVDLIKLVPGPLTPK-DSHWHHTA
MUL_3149|M.ulcerans_Agy99 PVTDRLGLDQMSWATPARWGFATSASTVDLIKLVPGPLTPK-DSHWHHTA
MAB_3698|M.abscessus_ATCC_1997 PVTGRAGLEQLSFLFPSRWGFAATAATCDLRKLVP--VGPQ-DELWNHDA
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAV_4325|M.avium_104 HIFVFDMAMLALLSIGYTGFVRWHIRLKR--
MMAR_1174|M.marinum_M SAWLFDMAMLAVLSIAYTAIVWWKIRLKRR-
MSMEG_1642|M.smegmatis_MC2_155 GTWLFDMAMLGVLSVFYVTFVRWKIRLKAG-
TH_4260|M.thermoresistible__bu GTWLFDMAMLAVLSVFYLALVRWKIRLPRA-
Mflv_4882|M.gilvum_PYR-GCK GTWLFDMGMLALISVGYLALVRWRIRLKSAG
Mvan_1546|M.vanbaalenii_PYR-1 GAWLFDMGMLLAISVGYLAFVRWRIRLKSA-
Mb1776|M.bovis_AF2122/97 SAWWFDMAMLVALSVIYVGFVRWKIRLKAC-
Rv1747|M.tuberculosis_H37Rv SAWWFDMAMLVALSVIYVGFVRWKIRLKAC-
MUL_3149|M.ulcerans_Agy99 SAWWFDMAMLAFISLCYVSFVRWKIRLKGG-
MAB_3698|M.abscessus_ATCC_1997 GTWLFDMGILGLLSLVMAGFVRWRIRLKG--
MLBr_00669|M.leprae_Br4923 -------------------------------