For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRPGSPALTVRYDGSTHTFAAGNDVVVGRDLRADVRIAHPLISRAHLVLRFDQGRWIAIDNGSLNGMYV NGRRVPAADISDGGQLHIGNPDGPLLTFEVGRHQGSAGTPPSTTAVPVPGRTSATWPAQHSGPTGRPPVQ RPPTGPYPGAGAYPPGGPRPVPAPPPPAAQSRPAHVQQPISAPAMDPVTVMGPAAAPRSSGSDGNIATSM LRILRPGRPADVPAGSIKIGRSSDNDIVIPDVLASRHHATLVPTPAGTEIRDNRSINGTFVNGSRVDSAI LADGDTVTIGNVDLSFRDGTLVRRTETAAATATGGLDVHGVTWTIENNKTLLDNISMSARPGTLTAVIGP SGAGKSTFARLVAGYTHPTTGQVSFEGHDVHAEYASLRSRIGMVPQDDVVHGQLTVRQALMYAAELRLPP DTTKADREQVVNEVLEELEMTKHLDTRVDKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVM TMLRQLADAGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGSAMGTTNWADIFSSVAGDPEAA AARYLSQNGPPPPQPPALTPSELGNPTRTSLRRQLSTIARRQIRLVISDRGYFAFLMLLPFIMGILSLSV PGDVGFGVPKTALEGGEAPNEPGQILVMLNVGAIFMGTALTIRALIGERAIFRREQAVGLSTSAYLLAKV VVFTAFAIVQAAIVTTIAIIGKGWGPGAVDSGAVVGDRSIELFLDIAVTCVASAMVGLALSALAKSAEQI MPLLVVAIMSQLVFSGGMIPVTDRLVLDQMSWFTPARWGFAASASTVDLIRLVPGPLTPQDRHWEHTPGT WLFDMGMLALISVGYLALVRWRIRLKSAG
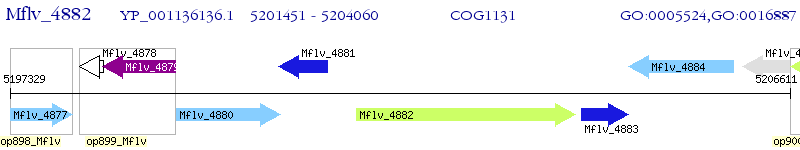
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4882 | - | - | 100% (869) | ABC transporter related |
| M. gilvum PYR-GCK | Mflv_2856 | - | 5e-25 | 37.84% (222) | ABC transporter related |
| M. gilvum PYR-GCK | Mflv_2197 | - | 3e-24 | 33.49% (215) | ABC transporter related |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1776 | - | 0.0 | 71.12% (883) | transmembrane ATP-binding protein ABC transporter |
| M. tuberculosis H37Rv | Rv1747 | - | 0.0 | 71.12% (883) | transmembrane ATP-binding protein ABC transporter |
| M. leprae Br4923 | MLBr_00669 | ftsE | 2e-22 | 34.57% (188) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_3698 | - | 0.0 | 56.28% (876) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_2612 | - | 0.0 | 72.79% (882) | transmembrane ATP-binding protein ABC transporter |
| M. avium 104 | MAV_3010 | - | 0.0 | 73.26% (875) | ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_1642 | - | 0.0 | 73.56% (904) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4260 | - | 0.0 | 81.71% (667) | PUTATIVE ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_3149 | - | 0.0 | 72.45% (882) | transmembrane ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_1546 | - | 0.0 | 89.69% (873) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1776|M.bovis_AF2122/97 -----MPMSQPAAPPVLTVRYEGSERTFAAGHDVVVGRDLRADVRVAHPL
Rv1747|M.tuberculosis_H37Rv -----MPMSQPAAPPVLTVRYEGSERTFAAGHDVVVGRDLRADVRVAHPL
MMAR_2612|M.marinum_M -------MTQTAAPPVLTVRYNGAERTFAAGHDVVIGRDLRADVRVADPL
MUL_3149|M.ulcerans_Agy99 MPLQLVPMTQTAAPPVLTVRYNGAERTFAAGHDVVIGRDLRADVRVADPL
MAV_3010|M.avium_104 ------------------MRYDGAERTFAAGNDVVIGRDLRADLRVAHPL
Mflv_4882|M.gilvum_PYR-GCK --------MSRPGSPALTVRYDGSTHTFAAGNDVVVGRDLRADVRIAHPL
Mvan_1546|M.vanbaalenii_PYR-1 --------MSRPATPALTVRYDGSTRSFAAGNDVVVGRDLRADIRIAHPL
MSMEG_1642|M.smegmatis_MC2_155 --------MSRPSPPALTVRYEGSTRTFAPGSDVVIGRDLRADVRIAHPL
TH_4260|M.thermoresistible__bu --------------------------------------------------
MAB_3698|M.abscessus_ATCC_1997 --------MTGREAPTLTVVFDGDERKFAPGHDIHIGRDIHADVRITHPL
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 ISRAHLLLRFDQGRWVAIDNGSLNGLYLNNRRVPVVDIYDAQRVHIGNPD
Rv1747|M.tuberculosis_H37Rv ISRAHLLLRFDQGRWVAIDNGSLNGLYLNNRRVPVVDIYDAQRVHIGNPD
MMAR_2612|M.marinum_M ISRVHLLLRFDQGRWIAIDNGSLNGLYINNRRVPVVDIQDGQRVNIGNPD
MUL_3149|M.ulcerans_Agy99 ISRVHLLLRFDQGRWIAIDNGSLNGLYINNRRVPVVDIQDGQRVNIGNPD
MAV_3010|M.avium_104 ISRTHLIVRYEQGRWVAIDNGSLNGLYVNNRRVPMVDINDGTRVNMGNPD
Mflv_4882|M.gilvum_PYR-GCK ISRAHLVLRFDQGRWIAIDNGSLNGMYVNGRRVPAADISDGGQLHIGNPD
Mvan_1546|M.vanbaalenii_PYR-1 ISRAHLVLRFDQGRWLAIDNGSLNGMYVNGRRVPAAEITDGTQVHIGNPD
MSMEG_1642|M.smegmatis_MC2_155 ISRAHLVLRFDQGRWVAIDNGSLNGMYVNGRRVSSVDLQDGQVLNIGNPD
TH_4260|M.thermoresistible__bu --------------------------------------------------
MAB_3698|M.abscessus_ATCC_1997 VSRIHLIIRFVDGKWIAEDQRSLNGIFVNGRQVGWVEVRDDTVINLGDPG
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 GPALDFEVG--RHRGSAGRPPQTTSIRLPN-LSAGAWPTDGPPQ--TGTL
Rv1747|M.tuberculosis_H37Rv GPALDFEVG--RHRGSAGRPPQTTSIRLPN-LSAGAWPTDGPPQ--TGTL
MMAR_2612|M.marinum_M GPALDFGVGR-RQQGSVGRPPPTAVMQVPPPMPSGAWPSQAPPQ--TGSH
MUL_3149|M.ulcerans_Agy99 GPALDFGVGR-RQQGSVGRPPPTAVMQVPPPMPSGPWPSQAPPQ--TGSH
MAV_3010|M.avium_104 GPALSFEVG--RHQGSAGRPPLTTSIPIVS-TPSGPMTGAAQGQP-PGTQ
Mflv_4882|M.gilvum_PYR-GCK GPLLTFEVG--RHQGSAGTPPSTTAVPVPG-RTSATWPAQHSGPT-GRPP
Mvan_1546|M.vanbaalenii_PYR-1 GPLLVFEVG--RHQGSAGSPPPTAAVPVAG-RPSATWPSQP-------PP
MSMEG_1642|M.smegmatis_MC2_155 GPQLSFEVG--RHQGSAGRTP-TAAVPVAG-HTGTSWPTQAPTGGGWQQP
TH_4260|M.thermoresistible__bu --------------------------------------------------
MAB_3698|M.abscessus_ATCC_1997 GPRLTFALGGLGGETQLGIAPPTMRGYVSGAQQQAPYTGAQQHGT-TGQP
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 GSGQLQQLPPATTRIPAAPPSGP--------QPRYPTGGQQLWPP-----
Rv1747|M.tuberculosis_H37Rv GSGQLQQLPPATTRIPAAPPSGP--------QPRYPTGGQQLWPP-----
MMAR_2612|M.marinum_M RPVPPQPAPP-TTRMPAQPPSGPQHVQPSAQQPQYPSGPQTSRSQ-----
MUL_3149|M.ulcerans_Agy99 RPVPPQPAPP-TTRMPAQPPSGPQHVQPSDQQPQYPSGPQTSRSQ-----
MAV_3010|M.avium_104 RISQPQQPASSSFRQPVQSPTGA--------QPSHPPSGPTRYPS-----
Mflv_4882|M.gilvum_PYR-GCK VQRPPTGPYPGAG--------------------AYPPGGPRPVPAPPPP-
Mvan_1546|M.vanbaalenii_PYR-1 SQPPPTGRPPAQR--------------------PYP-SAPRPSAPPTYP-
MSMEG_1642|M.smegmatis_MC2_155 YTQPPRTQYPQTTGTQQRYPSAPQHGYPNGPQTGYPSGPQRGYPSGPQTG
TH_4260|M.thermoresistible__bu --------------------------------------------------
MAB_3698|M.abscessus_ATCC_1997 YHSTQQHYPTGGYQSGPQTYGQP-------PSQQHPYGAAPSGPQTYG--
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 --------SGPQRAPQIYRPPT--AAP-----PPAG----ARGGTE-AGN
Rv1747|M.tuberculosis_H37Rv --------SGPQRAPQIYRPPT--AAP-----PPAG----ARGGTE-AGN
MMAR_2612|M.marinum_M --------TGAQPAPQIYRPPA--ASP-----APPAPLVVGRAPGE-TGN
MUL_3149|M.ulcerans_Agy99 --------TGAQPAPQIYRPPA--ASP-----APPAPLVVGRAPGE-TGN
MAV_3010|M.avium_104 --------SGLQHAPQIYRAPSGHAAPPT--TAQPAPAQTGRIGGD-SSN
Mflv_4882|M.gilvum_PYR-GCK --------AAQ-SRPAHVQQPISAPAMDP--VTVMGPAAAPRSSGS-DGN
Mvan_1546|M.vanbaalenii_PYR-1 --------TPQPSRPAPVQPPLSSPAMES--ATVMGPAAAPRSGG--DGN
MSMEG_1642|M.smegmatis_MC2_155 YPTNGPAGAPQSYQSQPVRTPPPAPANSSQAPTTMGPAAAPRGGAEPASN
TH_4260|M.thermoresistible__bu -----------------VRSPE-------------------------INN
MAB_3698|M.abscessus_ATCC_1997 --------TSGPTRPRQQYPSEPTPVP-------------ASYSNEPVEP
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 LATSMMKILRPGRLTGELPPGAVRIGRANDNDIVIPEVLASRHHATLVPT
Rv1747|M.tuberculosis_H37Rv LATSMMKILRPGRLTGELPPGAVRIGRANDNDIVIPEVLASRHHATLVPT
MMAR_2612|M.marinum_M IATSMMKILRPGRAAGELPPGAVTIGRADDNDIVIPEVLASRHHATLIPT
MUL_3149|M.ulcerans_Agy99 IATSMMKILRPGRAAGELPPGAVTIGRADDNDIVIPEVLASRHHATLIPT
MAV_3010|M.avium_104 LATSMMKILRPGRAGLESTPGAVKIGRANDNDIVIPEVLASRHHATLIPT
Mflv_4882|M.gilvum_PYR-GCK IATSMLRILRPGRP-ADVPAGSIKIGRSSDNDIVIPDVLASRHHATLVPT
Mvan_1546|M.vanbaalenii_PYR-1 IATSMLKILRPGRP-AEAPPGSIKIGRSTDNDIVIPDVLASRHHATLVHS
MSMEG_1642|M.smegmatis_MC2_155 LATSMLKILRPGRS-APAPAGAVKIGRATDNDIVIPDVLASRHHATLIPL
TH_4260|M.thermoresistible__bu LATSMMKILRPGRA-QEAPPGSIKIGRAVDNDIVIPDVLASRHHATLVPT
MAB_3698|M.abscessus_ATCC_1997 FGTRAMRALGIGGAPKPTP-GAITVGRAPDNTIVVNDVLASRHHALLLPT
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 PGGTEIRDNRSINGTFVNGARVDAALLHDGDVVTIGNIDLVFADGTLARR
Rv1747|M.tuberculosis_H37Rv PGGTEIRDNRSINGTFVNGARVDAALLHDGDVVTIGNIDLVFADGTLARR
MMAR_2612|M.marinum_M PQGTEIRDNRSINGTFVNGARVEAALLHDGDVVTIGNIDLIFANGALARR
MUL_3149|M.ulcerans_Agy99 PQGTEIRDNRSINGTFVNGARVEAALLHDGDVVTIGNIDLIFANGALARR
MAV_3010|M.avium_104 PAGTEIHDNRSINGTFVNGVRVDSAVLRDGDVVTIGNIDLVFHGGTLARR
Mflv_4882|M.gilvum_PYR-GCK PAGTEIRDNRSINGTFVNGSRVDSAILADGDTVTIGNVDLSFRDGTLVRR
Mvan_1546|M.vanbaalenii_PYR-1 PAGTEIRDNRSINGTFVNGARVDSAMLTEGDTVTIGNVDLVFRDGTLVRR
MSMEG_1642|M.smegmatis_MC2_155 PGGTEIRDERSINGTFVNGTRVDSAVLHDGDVVTIGNVDLVFSGGTLARR
TH_4260|M.thermoresistible__bu PAGMEIRDNRSINGTFVNGTRTETAVLNEGDVVTIGNVDLVFRNGTLVRR
MAB_3698|M.abscessus_ATCC_1997 PGGLEIQDLHTINGTFVNGVQVEQSSLREGDTVTIGNVDLVVQDGTLVKP
MLBr_00669|M.leprae_Br4923 -------------------------------MITLDHVSKKYKS------
:*:.::. .
Mb1776|M.bovis_AF2122/97 EENLLETRVGGLDVRGVTWTIDGDKTLLDGISLTARPGMLTAVIGPSGAG
Rv1747|M.tuberculosis_H37Rv EENLLETRVGGLDVRGVTWTIDGDKTLLDGISLTARPGMLTAVIGPSGAG
MMAR_2612|M.marinum_M EESLLETRTGGLDVHGVTWTIEGNKTLLDNISLAARPGMLTAVIGPSGAG
MUL_3149|M.ulcerans_Agy99 EESLLETRTGGLDVHGVTWTIEGNKTLLDNISLAARPGMLTAVIGPSGAG
MAV_3010|M.avium_104 DQTAAATRTGGLDVRGVTWTIENNKTLLDNISLTALPGTLTAIIGPSGAG
Mflv_4882|M.gilvum_PYR-GCK TETAAATATGGLDVHGVTWTIENNKTLLDNISMSARPGTLTAVIGPSGAG
Mvan_1546|M.vanbaalenii_PYR-1 TETAAATATGGLDVHGVTWTIENNKTLLDNISMSARPGTLTAVIGPSGAG
MSMEG_1642|M.smegmatis_MC2_155 SETEADTRTGGLEVRGLTWTIEGNKTLLDNISIDARPGTLTAVIGPSGAG
TH_4260|M.thermoresistible__bu TETEAATDAGGLDVRGVTWTIENNKTLLDNISITARPGTLTAIIGPSGAG
MAB_3698|M.abscessus_ATCC_1997 T---TATRVGGLEVQSIDFVVEGGKKLLDDISFSAGPGSLTAVIGPSGAG
MLBr_00669|M.leprae_Br4923 ----------------------LARPALDNVNVKIDKGEFVFLIGPSGSG
: **.:.. * :. :*****:*
Mb1776|M.bovis_AF2122/97 KSTLARLVAGYTHPTDGTVTFEGHNVHAEY----ASLRSRIGMVPQDDVV
Rv1747|M.tuberculosis_H37Rv KSTLARLVAGYTHPTDGTVTFEGHNVHAEY----ASLRSRIGMVPQDDVV
MMAR_2612|M.marinum_M KSTFARLVAGYTHPTNGTVTFEGHNIHAEY----ASLRSRIGMVPQDDVV
MUL_3149|M.ulcerans_Agy99 KSTFARLVAGYTHPTNGTVTFEGHNIHAEY----ASLRSRIGMVPQDDVV
MAV_3010|M.avium_104 KSTFARLVAGYTHPTTGTVAFEGHNVHAEY----ASLRSRIGMVPQDDVV
Mflv_4882|M.gilvum_PYR-GCK KSTFARLVAGYTHPTTGQVSFEGHDVHAEY----ASLRSRIGMVPQDDVV
Mvan_1546|M.vanbaalenii_PYR-1 KSTFARLVAGYTHPTTGQVSFEGHDVHAEY----ASLRSRIGMVPQDDVV
MSMEG_1642|M.smegmatis_MC2_155 KSTFARQVAGYTHPTSGTIKFEGHDVHAEY----ASLRSRIGMVPQDDVV
TH_4260|M.thermoresistible__bu KSTFARLIAGYTHPTSGTVTFEGHNVHAEY----ASLRSRIGMVPQDDVV
MAB_3698|M.abscessus_ATCC_1997 KSTLARLIVGNTQPSAGKVSFEGHGVHAEY----AVMRNRIGMVPQDDVV
MLBr_00669|M.leprae_Br4923 KSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRLRQVIGCVFQDFRL
***: * : * *: * : .. . :: . :*. ** * ** :
Mb1776|M.bovis_AF2122/97 HGQLTVKHALMYAAELRLPPDTTKDDRTQVVARVLEELEMSKHIDTRVDK
Rv1747|M.tuberculosis_H37Rv HGQLTVKHALMYAAELRLPPDTTKDDRTQVVARVLEELEMSKHIDTRVDK
MMAR_2612|M.marinum_M HGQLTVRQALMYAAELRLPPDTTKEDREQVVARVLEELEMSKHLETRVDK
MUL_3149|M.ulcerans_Agy99 HGQLTVRQALMYAAELRLPPDTTKEDREHVVARVLEELEMSKHLETRVDK
MAV_3010|M.avium_104 HGQLTVQQALMYAAELRLPPDTTKEDRAQVVARVLEELEMTQHLHTRVDK
Mflv_4882|M.gilvum_PYR-GCK HGQLTVRQALMYAAELRLPPDTTKADREQVVNEVLEELEMTKHLDTRVDK
Mvan_1546|M.vanbaalenii_PYR-1 HGQLTVRQALMYAAELRLPPDTTKADREQVVNEVLEELEMTKHLDTRVDK
MSMEG_1642|M.smegmatis_MC2_155 HGQLTVRQALMYAAELRLPPDTTKEDREQVVMQVLEELEMTKHLDTRVDK
TH_4260|M.thermoresistible__bu HGQLTVNQALMYAAELRLPPDTTEEDRRQVVAEVLEELEMTAHADTRVDK
MAB_3698|M.abscessus_ATCC_1997 HPQLTVSQALHYAAELRLPPDTTKEDREQVVSRVLEELEMTKHAETRVDK
MLBr_00669|M.leprae_Br4923 LQQKTVYENVAFALEVIG---RRSDAINQVVPDVLETVGLSGKANRLPDE
* ** . : :* *: . :** *** : :: : . *:
Mb1776|M.bovis_AF2122/97 LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
Rv1747|M.tuberculosis_H37Rv LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
MMAR_2612|M.marinum_M LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
MUL_3149|M.ulcerans_Agy99 LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
MAV_3010|M.avium_104 LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
Mflv_4882|M.gilvum_PYR-GCK LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
Mvan_1546|M.vanbaalenii_PYR-1 LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
MSMEG_1642|M.smegmatis_MC2_155 LSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
TH_4260|M.thermoresistible__bu LSGGQRKRASVAMELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLADAG
MAB_3698|M.abscessus_ATCC_1997 LSGGQRKRASVAMELLTGPSLLILDEPTSGLDPALDRQVMSLMRQLADAG
MLBr_00669|M.leprae_Br4923 LSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERINRTG
****:::*.::* ::. * :*: ****..*** .:::* ::.:: :*
Mb1776|M.bovis_AF2122/97 RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPTQIGPVMGTTNWADIF
Rv1747|M.tuberculosis_H37Rv RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPTQIGPVMGTTNWADIF
MMAR_2612|M.marinum_M RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPSQIGPAMGTTNWADIF
MUL_3149|M.ulcerans_Agy99 RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPSQIGPAMGTTNWADIF
MAV_3010|M.avium_104 RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPGQIGPAMGTTNWADIF
Mflv_4882|M.gilvum_PYR-GCK RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGSAMGTTNWADIF
Mvan_1546|M.vanbaalenii_PYR-1 RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGPSMGTTNWADIF
MSMEG_1642|M.smegmatis_MC2_155 RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFFGPPSQIGPELGTTNWADIF
TH_4260|M.thermoresistible__bu RVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGPTMGTTNWADIF
MAB_3698|M.abscessus_ATCC_1997 RVVVVVTHSLTYLSYCDQVLLLAPGGKTAYCGPPRSIGQEMGTTDWADIF
MLBr_00669|M.leprae_Br4923 TTVLMATHDHHIVDSMRQRVVELSLG-------------RLVRDEWCGIY
.*::.**. :. * :: . * : :*..*:
Mb1776|M.bovis_AF2122/97 STVADDPDAAKARYLARTGPTPPPPP-VEQPAELG-DPAHTSLFRQFSTI
Rv1747|M.tuberculosis_H37Rv STVADDPDAAKARYLARTGPTPPPPP-VEQPAELG-DPAHTSLFRQFSTI
MMAR_2612|M.marinum_M STVADDPEGAQARYLAQTGPPPPAPP-VERPGELGGDPSHTSLFRQFSTI
MUL_3149|M.ulcerans_Agy99 STVADDPEGAQARYLAQTGPPPPAPP-VERPGELGGDPSHTSLFRQFSTI
MAV_3010|M.avium_104 STVAGDPDGSRARYLARTGPQPPPPP-AEQPAELG-DPAHTSLVRQFSTI
Mflv_4882|M.gilvum_PYR-GCK SSVAGDPEAAAARYLSQNGPPPPQPP-ALTPSELG-NPTRTSLRRQLSTI
Mvan_1546|M.vanbaalenii_PYR-1 SAVAGDPDAAAQRYLAQNGPPPPQPP-ALTPSELG-NPTRTSLRRQLSTI
MSMEG_1642|M.smegmatis_MC2_155 STVADDPAEANRRYLARTPEAPPASASSEAPADLG-APAKTSLRRQFSTI
TH_4260|M.thermoresistible__bu STVAGDPDAANRRFLERQGPPRELPAESAKPTDLG-SPTRTSLRRQFSTI
MAB_3698|M.abscessus_ATCC_1997 ANAAADPDGVHQAYLSRHPAANKPATAASPPGDLG-KPPKTSLLRQVSTV
MLBr_00669|M.leprae_Br4923 GMDR----------------------------------------------
.
Mb1776|M.bovis_AF2122/97 ARRQLRLIVSDRGYFVFLALLPFIMGALSMSVPGDVGFGFPN---PMGDA
Rv1747|M.tuberculosis_H37Rv ARRQLRLIVSDRGYFVFLALLPFIMGALSMSVPGDVGFGFPN---PMGDA
MMAR_2612|M.marinum_M ARRQMRLIISDRGYFIFLAILPFIMGSLSMSVPGDVGFGMPN---PMGNA
MUL_3149|M.ulcerans_Agy99 ARRQMRLIISDRGYFIFLAILPFVMGSLSMSVPGDVGFGMPN---PMGNA
MAV_3010|M.avium_104 ARRQMRLIVSDRGYFVFLALLPFIMGSLSMSVPGDVGFGIPN---PMGAA
Mflv_4882|M.gilvum_PYR-GCK ARRQIRLVISDRGYFAFLMLLPFIMGILSLSVPGDVGFGVPKTALEGGEA
Mvan_1546|M.vanbaalenii_PYR-1 ARRQIRLVISDRGYFAFLMLLPFIMGVLSLSVPGDVGFGVPKTALEGGEA
MSMEG_1642|M.smegmatis_MC2_155 GRRQLRLIISDRGYFIFLALLPFIMGTLSLSVPGTVGFGVPN---PMGDA
TH_4260|M.thermoresistible__bu ARRQIRLIVSDRGYFVFLALLPFIMGVLSLSVPGDVGFGVPN---PMGDA
MAB_3698|M.abscessus_ATCC_1997 ARRQGRLILADRGYFIFLALLPFVLAGLVLSVPGSAGFNVALP-----DN
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 PNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLSTTAYLIA
Rv1747|M.tuberculosis_H37Rv PNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLSTTAYLIA
MMAR_2612|M.marinum_M PNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLSTTAYLLA
MUL_3149|M.ulcerans_Agy99 PNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLSTTAYLLA
MAV_3010|M.avium_104 PNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFLREQAVGLSTSAYLLA
Mflv_4882|M.gilvum_PYR-GCK PNEPGQILVMLNVGAIFMGTALTIRALIGERAIFRREQAVGLSTSAYLLA
Mvan_1546|M.vanbaalenii_PYR-1 PNEPGQILVMLNVGAIFMGTALTIRALISERAIFRREQAVGLSTSAYLLA
MSMEG_1642|M.smegmatis_MC2_155 PNEPGQILVLLNVGAIFMGTALTIRDLIGERAIFRREQAVGLSTTAYLLA
TH_4260|M.thermoresistible__bu PNEPGQILVLLNVGAIFMGTALTIRDLIGERAIFRREQAVGLSTTAYLLA
MAB_3698|M.abscessus_ATCC_1997 PNEPGLLLALTNLGASFLGLALTIRDLIGERVIFRREQAVGLSATAYLLA
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 KVCVYTVLAVVQSAIVTVIVLVGKGGP-----TQGAVALSKPDLELFVDV
Rv1747|M.tuberculosis_H37Rv KVCVYTVLAVVQSAIVTVIVLVGKGGP-----TQGAVALSKPDLELFVDV
MMAR_2612|M.marinum_M KICVYTVFAVVQSAIVTIIVLIGKGGP-----TQGAVALGKPGLELFADV
MUL_3149|M.ulcerans_Agy99 KICVYTVFAVVQSAIVTIIVLIGKGGP-----TQGAVALGKPGLELFADV
MAV_3010|M.avium_104 KVCVYTVFAIVQSAIVTTIAVLGKGAP-----TQGAVALGKPALELFADV
Mflv_4882|M.gilvum_PYR-GCK KVVVFTAFAIVQAAIVTTIAIIGKGWG--PGAVDSGAVVGDRSIELFLDI
Mvan_1546|M.vanbaalenii_PYR-1 KVVVFTAFAIVQAAIVTSIAIAGKGWG--PGAVERGAVIGNRSVELFTDI
MSMEG_1642|M.smegmatis_MC2_155 KVCVYSVFAVVQSAIVTVITISGKGWG--EGAVERGVVIPNRSLELFLSM
TH_4260|M.thermoresistible__bu KVCVYSVFAIVQSAIVTAITIAGKGWDVDKGAVDHGSVFGNPKLELFFGM
MAB_3698|M.abscessus_ATCC_1997 KIITYCGAAIVQSAIMTAVIVIVRGGP-----TQGAVLLGSANFELWVAV
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 AVTCVASAMLGLALSAIAKSNEQIMPLLVVAVMSQLVFSGGMIPVTGRVP
Rv1747|M.tuberculosis_H37Rv AVTCVASAMLGLALSAIAKSNEQIMPLLVVAVMSQLVFSGGMIPVTGRVP
MMAR_2612|M.marinum_M AATCVASAMLGLALSAVAKSNEQIMPLLVVAVMSQLVFSGGMIPVTDRLG
MUL_3149|M.ulcerans_Agy99 AATCVASAMLGLALSAVAKSNEQIMPLLVVAVMSQLVFSGGMIPVTDRLG
MAV_3010|M.avium_104 ALTCVASAMLGLALSAVAKSNEQIMPLLVVAVMSQLVFSGGMIPVTGRVG
Mflv_4882|M.gilvum_PYR-GCK AVTCVASAMVGLALSALAKSAEQIMPLLVVAIMSQLVFSGGMIPVTDRLV
Mvan_1546|M.vanbaalenii_PYR-1 AVTCVAAAMVGLALSALAKSAEQIMPLLVVAIMSQLVFSGGMIPVTDRIV
MSMEG_1642|M.smegmatis_MC2_155 AATTVTAAMVGLALSALAKTSEQIMPLLVVAIMSQLVFSGGMIPVTGRIG
TH_4260|M.thermoresistible__bu AATTVAAAMLGLLLSALAKTSEQIMPLLVVAIMSQLVFSGGMIPVTDRIG
MAB_3698|M.abscessus_ATCC_1997 AATACAATVLGLVLSSLATSAEQVMPMLVIATMTQIVFSGGLIPVTGRAG
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 LDQMSWVTPARWGFAASAATVDLIKLVPGPLTPKDSHWHHTASAWWFDMA
Rv1747|M.tuberculosis_H37Rv LDQMSWVTPARWGFAASAATVDLIKLVPGPLTPKDSHWHHTASAWWFDMA
MMAR_2612|M.marinum_M LDQMSWATPARWGFATSASTVDLIKLVPGPLTPKDSHWHHTAGAWWFDMA
MUL_3149|M.ulcerans_Agy99 LDQMSWATPARWGFATSASTVDLIKLVPGPLTPKDSHWHHTASAWWFDMA
MAV_3010|M.avium_104 LDQMAWATPARWGFAASASTADLTKLVPGPLTPKDSHWRHAPGTWWFDVG
Mflv_4882|M.gilvum_PYR-GCK LDQMSWFTPARWGFAASASTVDLIRLVPGPLTPQDRHWEHTPGTWLFDMG
Mvan_1546|M.vanbaalenii_PYR-1 LDQMSWFTPARWGFAASASTVDLIRLVPGPLTPQDRHWQHTTGAWLFDMG
MSMEG_1642|M.smegmatis_MC2_155 LDQLSWITPARWGFAASASTVDLIRLVPGPLTPKDSHWEHTSGTWLFDMA
TH_4260|M.thermoresistible__bu LDQLSWFTPARWGFAASASTADLITLVPGPLTPKDRHWEHDSGTWLFDMA
MAB_3698|M.abscessus_ATCC_1997 LEQLSFLFPSRWGFAATAATCDLRKLVP--VGPQDELWNHDAGTWLFDMG
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1776|M.bovis_AF2122/97 MLVALSVIYVGFVRWKIRLKAC-
Rv1747|M.tuberculosis_H37Rv MLVALSVIYVGFVRWKIRLKAC-
MMAR_2612|M.marinum_M MLAFISLCYVSFVRWKIRLKGG-
MUL_3149|M.ulcerans_Agy99 MLAFISLCYVSFVRWKIRLKGG-
MAV_3010|M.avium_104 MLILISTVYLGFVRWKIRLKGG-
Mflv_4882|M.gilvum_PYR-GCK MLALISVGYLALVRWRIRLKSAG
Mvan_1546|M.vanbaalenii_PYR-1 MLLAISVGYLAFVRWRIRLKSA-
MSMEG_1642|M.smegmatis_MC2_155 MLGVLSVFYVTFVRWKIRLKAG-
TH_4260|M.thermoresistible__bu MLAVLSVFYLALVRWKIRLPRA-
MAB_3698|M.abscessus_ATCC_1997 ILGLLSLVMAGFVRWRIRLKG--
MLBr_00669|M.leprae_Br4923 -----------------------