For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVVGRHVQADVRIPDPRISRAHLILRFEQGRWLAIDNGSLNGTYLNGYRMPVVDIHDGQSIHVGNPHGPR LTFALGPQQGHAGHPGRPRPSHDSGPPTLTWTAIAEQTNPTRPPRRPVPPRRPARPPSTPPEERTVIVPG SPARSRVGPASAPAGPPRTSPTPATEFTVIDAKTADVSNLATRFVKLLSPRASSAAGTAGAMTIGRAADN DIVVADVLASRHHATLVPTALGTEIRDNSVNGTFVNGTRVGSAILSDDDVVTIGNVDLVFRDGTLTERGQ TATRSGGLEVRNVTYVVDEGKQLLDDISLTARPGTLTAVIGGSGAGKSTLARLIAGYTTPSSGSVTFEGH NIHGEYASLRSRIGMVPQDDVVHRQLTVNQALGYAAELRLPPDTGKADRARVVAQVLEELDLTKHADTRV DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDLQVMTMLRQLADAGRVVLVVTHSLSYLDVCDQ VLLMAPGGKTAYCGAPDRIGPAMGTTNWAKIFTQVGADPEEANRRFREQREAQQRSHKSDRADEPEDLGE PVHTSLRHQISTIARRQVRLVVADRAYFVFLALLPFILGSLSLTVPGHNGFRVPAPAAGTPDEAAQILAL LLPAAAFMGTALTIRDLVGERAIFQREQAVGLSTTAYLLAKTAVFCGFAVLQSAIVTAIVVIGKGAPTRG AVLFGHSTAGAVAELFATVAATCVASAVLGLAISSLVRSSEQIMPLFVVTVMAQLVLCGGMVPVTGRLGL NQLSWAMPARWGYAAASATVDLRHLVPDSLLSQDRFWLHTPGTWLLDMGMLAALALCYAVFVRWKIRLRR
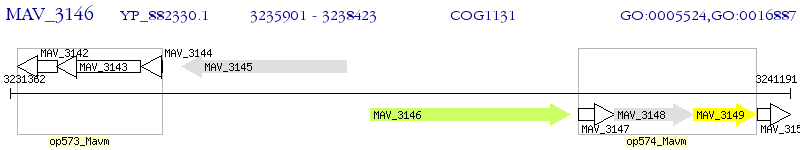
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3146 | - | - | 100% (840) | ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_4325 | - | 0.0 | 63.01% (857) | ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_3010 | - | 0.0 | 61.73% (857) | ABC transporter, ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1776 | - | 0.0 | 62.13% (853) | transmembrane ATP-binding protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_4882 | - | 0.0 | 62.21% (852) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1747 | - | 0.0 | 62.13% (853) | transmembrane ATP-binding protein ABC transporter |
| M. leprae Br4923 | MLBr_00669 | ftsE | 3e-22 | 33.33% (210) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_3698 | - | 0.0 | 55.46% (851) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_1174 | - | 0.0 | 62.86% (859) | transmembrane ATP-binding protein ABC transporter |
| M. smegmatis MC2 155 | MSMEG_1642 | - | 0.0 | 60.96% (876) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4260 | - | 0.0 | 68.30% (672) | PUTATIVE ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_3149 | - | 0.0 | 62.35% (858) | transmembrane ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_1546 | - | 0.0 | 61.01% (854) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1642|M.smegmatis_MC2_155 -------MSRPSPP-ALTVRYEGSTRTFAPGSDVVIGRDLRADVRIAHPL
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK -------MSRPGSP-ALTVRYDGSTHTFAAGNDVVVGRDLRADVRIAHPL
Mvan_1546|M.vanbaalenii_PYR-1 -------MSRPATP-ALTVRYDGSTRSFAAGNDVVVGRDLRADIRIAHPL
Mb1776|M.bovis_AF2122/97 -----MPMSQPAAPPVLTVRYEGSERTFAAGHDVVVGRDLRADVRVAHPL
Rv1747|M.tuberculosis_H37Rv -----MPMSQPAAPPVLTVRYEGSERTFAAGHDVVVGRDLRADVRVAHPL
MUL_3149|M.ulcerans_Agy99 MPLQLVPMTQTAAPPVLTVRYNGAERTFAAGHDVVIGRDLRADVRVADPL
MMAR_1174|M.marinum_M -------MPQPAPP-ALTVRCDGSERTFAAGHDVVVGRDLRADMRITHPL
MAV_3146|M.avium_104 ---------------------------------MVVGRHVQADVRIPDPR
MAB_3698|M.abscessus_ATCC_1997 -------MTGREAP-TLTVVFDGDERKFAPGHDIHIGRDIHADVRITHPL
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 ISRAHLVLRFDQGRWVAIDNGSLNGMYVNGRRVSSVDLQDGQVLNIGNPD
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK ISRAHLVLRFDQGRWIAIDNGSLNGMYVNGRRVPAADISDGGQLHIGNPD
Mvan_1546|M.vanbaalenii_PYR-1 ISRAHLVLRFDQGRWLAIDNGSLNGMYVNGRRVPAAEITDGTQVHIGNPD
Mb1776|M.bovis_AF2122/97 ISRAHLLLRFDQGRWVAIDNGSLNGLYLNNRRVPVVDIYDAQRVHIGNPD
Rv1747|M.tuberculosis_H37Rv ISRAHLLLRFDQGRWVAIDNGSLNGLYLNNRRVPVVDIYDAQRVHIGNPD
MUL_3149|M.ulcerans_Agy99 ISRVHLLLRFDQGRWIAIDNGSLNGLYINNRRVPVVDIQDGQRVNIGNPD
MMAR_1174|M.marinum_M ISRAHLLLRFDQGRWLAIDNNSLNGTFVNGRRVPMVEIYDGQSLNIGNPD
MAV_3146|M.avium_104 ISRAHLILRFEQGRWLAIDNGSLNGTYLNGYRMPVVDIHDGQSIHVGNPH
MAB_3698|M.abscessus_ATCC_1997 VSRIHLIIRFVDGKWIAEDQRSLNGIFVNGRQVGWVEVRDDTVINLGDPG
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 GPQLSFEVG--RHQGSAGRTP-TAAVPVAG-HTGTSWPTQAPTGGGWQQP
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK GPLLTFEVG--RHQGSAGTPPSTTAVPVPG-RTSATWPAQHS-GPTGRPP
Mvan_1546|M.vanbaalenii_PYR-1 GPLLVFEVG--RHQGSAGSPPPTAAVPVAG-RPSATWPSQP-------PP
Mb1776|M.bovis_AF2122/97 GPALDFEVG--RHRGSAGRPPQTTSIRLPN-LSAGAWPTDGP--PQTGTL
Rv1747|M.tuberculosis_H37Rv GPALDFEVG--RHRGSAGRPPQTTSIRLPN-LSAGAWPTDGP--PQTGTL
MUL_3149|M.ulcerans_Agy99 GPALDFGVGR-RQQGSVGRPPPTAVMQVPPPMPSGPWPSQAP--PQTGSH
MMAR_1174|M.marinum_M GPLLSFEVG--RHVGMAGRPPQTESMRVTP--SMVRPSGQHPVAHGPTST
MAV_3146|M.avium_104 GPRLTFALG--PQQGHAGHPGRPRPSHDSG-PPTLTWTAIAE----QTNP
MAB_3698|M.abscessus_ATCC_1997 GPRLTFALGGLGGETQLGIAPPTMRGYVSGAQQQAPYTGAQQHGTTGQPY
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 YTQPPRTQYPQTTGTQQRYPSAPQHGYPNGPQTGYPSGPQRGYPSGPQTG
TH_4260|M.thermoresistible__bu --------------------------------------------------
Mflv_4882|M.gilvum_PYR-GCK VQRPPTGPYPGAGAYPPGGPRPVPAPPPPAAQ-SRPAHVQQPISAPAMDP
Mvan_1546|M.vanbaalenii_PYR-1 SQPPPTGRPPAQRPYP-SAPRPSAPPTYPTPQPSRPAPVQPPLSSPAMES
Mb1776|M.bovis_AF2122/97 GSGQLQQLPPATTRIPAAPPSGP--------QPRYPTGGQQLWPPSGPQR
Rv1747|M.tuberculosis_H37Rv GSGQLQQLPPATTRIPAAPPSGP--------QPRYPTGGQQLWPPSGPQR
MUL_3149|M.ulcerans_Agy99 RPVPPQPAPP-TTRMPAQPPSGPQHVQPSDQQPQYPSGPQTSRSQTGAQP
MMAR_1174|M.marinum_M GSHTSHEAPPTARFHPGAAPPGPARTGQPMYPSSSAPWRPTGYPASPAPP
MAV_3146|M.avium_104 TRPPRRPVPPRRPARPPSTPPEER----TVIVPGSPARSRVG-PASAPAG
MAB_3698|M.abscessus_ATCC_1997 HSTQQHYPTGGYQSGPQTYGQPPSQQHPYGAAPSGPQTYGTSGPTRPRQQ
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 YPTNG--PAGAPQSYQSQPVRTPPPAPANSSQAPTTMGPAAAPRGGAEPA
TH_4260|M.thermoresistible__bu -------------------VRSPE-------------------------I
Mflv_4882|M.gilvum_PYR-GCK VTVMG--PAAAP--------RSSGS------------------------D
Mvan_1546|M.vanbaalenii_PYR-1 ATVMG--PAAAP--------RSGG-------------------------D
Mb1776|M.bovis_AF2122/97 APQIYRPPTAAPPPAG----ARGGT-----------------------EA
Rv1747|M.tuberculosis_H37Rv APQIYRPPTAAPPPAG----ARGGT-----------------------EA
MUL_3149|M.ulcerans_Agy99 APQIYRPPAASPAPPAPLVVGRAPG-----------------------ET
MMAR_1174|M.marinum_M PPSNFPPPPPRPGPQPPQQATKPPE------------------------V
MAV_3146|M.avium_104 PPRTSPTPATEFTVID----AKTAD------------------------V
MAB_3698|M.abscessus_ATCC_1997 YPSEPTPVPASYSNEP---------------------------------V
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 SNLATSMLKILRPGRSA-PAPAGAVKIGRATDNDIVIPDVLASRHHATLI
TH_4260|M.thermoresistible__bu NNLATSMMKILRPGRAQ-EAPPGSIKIGRAVDNDIVIPDVLASRHHATLV
Mflv_4882|M.gilvum_PYR-GCK GNIATSMLRILRPGRPA-DVPAGSIKIGRSSDNDIVIPDVLASRHHATLV
Mvan_1546|M.vanbaalenii_PYR-1 GNIATSMLKILRPGRPA-EAPPGSIKIGRSTDNDIVIPDVLASRHHATLV
Mb1776|M.bovis_AF2122/97 GNLATSMMKILRPGRLTGELPPGAVRIGRANDNDIVIPEVLASRHHATLV
Rv1747|M.tuberculosis_H37Rv GNLATSMMKILRPGRLTGELPPGAVRIGRANDNDIVIPEVLASRHHATLV
MUL_3149|M.ulcerans_Agy99 GNIATSMMKILRPGRAAGELPPGAVTIGRADDNDIVIPEVLASRHHATLI
MMAR_1174|M.marinum_M ANLATKMFHALLPRSGGAERPAGSVTIGRATDNDIVIPDVLASRHHAFLT
MAV_3146|M.avium_104 SNLATRFVKLLSPRASSAAGTAGAMTIGRAADNDIVVADVLASRHHATLV
MAB_3698|M.abscessus_ATCC_1997 EPFGTRAMRALGIGGAP-KPTPGAITVGRAPDNTIVVNDVLASRHHALLL
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 PLPGGTEIRDERSINGTFVNGTRVDSAVLHDGDVVTIGNVDLVFSGGTLA
TH_4260|M.thermoresistible__bu PTPAGMEIRDNRSINGTFVNGTRTETAVLNEGDVVTIGNVDLVFRNGTLV
Mflv_4882|M.gilvum_PYR-GCK PTPAGTEIRDNRSINGTFVNGSRVDSAILADGDTVTIGNVDLSFRDGTLV
Mvan_1546|M.vanbaalenii_PYR-1 HSPAGTEIRDNRSINGTFVNGARVDSAMLTEGDTVTIGNVDLVFRDGTLV
Mb1776|M.bovis_AF2122/97 PTPGGTEIRDNRSINGTFVNGARVDAALLHDGDVVTIGNIDLVFADGTLA
Rv1747|M.tuberculosis_H37Rv PTPGGTEIRDNRSINGTFVNGARVDAALLHDGDVVTIGNIDLVFADGTLA
MUL_3149|M.ulcerans_Agy99 PTPQGTEIRDNRSINGTFVNGARVEAALLHDGDVVTIGNIDLIFANGALA
MMAR_1174|M.marinum_M LTPLGTEIRDAHSVNGTFVNGVRVGSAVLSDGDVVTIGNVDLVFTGGTLI
MAV_3146|M.avium_104 PTALGTEIRDN-SVNGTFVNGTRVGSAILSDDDVVTIGNVDLVFRDGTLT
MAB_3698|M.abscessus_ATCC_1997 PTPGGLEIQDLHTINGTFVNGVQVEQSSLREGDTVTIGNVDLVVQDGTLV
MLBr_00669|M.leprae_Br4923 ---------------------------------MITLDHVSKKYKS----
:*:.::. .
MSMEG_1642|M.smegmatis_MC2_155 RRSETEADTRTGGLEVRGLTWTIEGNKTLLDNISIDARPGTLTAVIGPSG
TH_4260|M.thermoresistible__bu RRTETEAATDAGGLDVRGVTWTIENNKTLLDNISITARPGTLTAIIGPSG
Mflv_4882|M.gilvum_PYR-GCK RRTETAAATATGGLDVHGVTWTIENNKTLLDNISMSARPGTLTAVIGPSG
Mvan_1546|M.vanbaalenii_PYR-1 RRTETAAATATGGLDVHGVTWTIENNKTLLDNISMSARPGTLTAVIGPSG
Mb1776|M.bovis_AF2122/97 RREENLLETRVGGLDVRGVTWTIDGDKTLLDGISLTARPGMLTAVIGPSG
Rv1747|M.tuberculosis_H37Rv RREENLLETRVGGLDVRGVTWTIDGDKTLLDGISLTARPGMLTAVIGPSG
MUL_3149|M.ulcerans_Agy99 RREESLLETRTGGLDVHGVTWTIEGNKTLLDNISLAARPGMLTAVIGPSG
MMAR_1174|M.marinum_M RR--TEAATRSGGLEVKSVCFTVDGGKQLLDHISLTARPGTLTAIIGGSG
MAV_3146|M.avium_104 ER--GQTATRSGGLEVRNVTYVVDEGKQLLDDISLTARPGTLTAVIGGSG
MAB_3698|M.abscessus_ATCC_1997 KP---TTATRVGGLEVQSIDFVVEGGKKLLDDISFSAGPGSLTAVIGPSG
MLBr_00669|M.leprae_Br4923 ------------------------LARPALDNVNVKIDKGEFVFLIGPSG
: ** :.. * :. :** **
MSMEG_1642|M.smegmatis_MC2_155 AGKSTFARQVAGYTHPTSGTIKFEGHDVHAEY----ASLRSRIGMVPQDD
TH_4260|M.thermoresistible__bu AGKSTFARLIAGYTHPTSGTVTFEGHNVHAEY----ASLRSRIGMVPQDD
Mflv_4882|M.gilvum_PYR-GCK AGKSTFARLVAGYTHPTTGQVSFEGHDVHAEY----ASLRSRIGMVPQDD
Mvan_1546|M.vanbaalenii_PYR-1 AGKSTFARLVAGYTHPTTGQVSFEGHDVHAEY----ASLRSRIGMVPQDD
Mb1776|M.bovis_AF2122/97 AGKSTLARLVAGYTHPTDGTVTFEGHNVHAEY----ASLRSRIGMVPQDD
Rv1747|M.tuberculosis_H37Rv AGKSTLARLVAGYTHPTDGTVTFEGHNVHAEY----ASLRSRIGMVPQDD
MUL_3149|M.ulcerans_Agy99 AGKSTFARLVAGYTHPTNGTVTFEGHNIHAEY----ASLRSRIGMVPQDD
MMAR_1174|M.marinum_M AGKTTLSRLIAGYTSPSSGTVTFEGHNIHAEY----ASMRSRIGMVPQDD
MAV_3146|M.avium_104 AGKSTLARLIAGYTTPSSGSVTFEGHNIHGEY----ASLRSRIGMVPQDD
MAB_3698|M.abscessus_ATCC_1997 AGKSTLARLIVGNTQPSAGKVSFEGHGVHAEY----AVMRNRIGMVPQDD
MLBr_00669|M.leprae_Br4923 SGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRLRQVIGCVFQDF
:**:*: * : * *: * : .. . :: . :*. ** * **
MSMEG_1642|M.smegmatis_MC2_155 VVHGQLTVRQALMYAAELRLPPDTTKEDREQVVMQVLEELEMTKHLDTRV
TH_4260|M.thermoresistible__bu VVHGQLTVNQALMYAAELRLPPDTTEEDRRQVVAEVLEELEMTAHADTRV
Mflv_4882|M.gilvum_PYR-GCK VVHGQLTVRQALMYAAELRLPPDTTKADREQVVNEVLEELEMTKHLDTRV
Mvan_1546|M.vanbaalenii_PYR-1 VVHGQLTVRQALMYAAELRLPPDTTKADREQVVNEVLEELEMTKHLDTRV
Mb1776|M.bovis_AF2122/97 VVHGQLTVKHALMYAAELRLPPDTTKDDRTQVVARVLEELEMSKHIDTRV
Rv1747|M.tuberculosis_H37Rv VVHGQLTVKHALMYAAELRLPPDTTKDDRTQVVARVLEELEMSKHIDTRV
MUL_3149|M.ulcerans_Agy99 VVHGQLTVRQALMYAAELRLPPDTTKEDREHVVARVLEELEMSKHLETRV
MMAR_1174|M.marinum_M VVHRQLTVSQALGYAAELRLPPDTSKEDRAQVVAQVLEELELTKHAETRV
MAV_3146|M.avium_104 VVHRQLTVNQALGYAAELRLPPDTGKADRARVVAQVLEELDLTKHADTRV
MAB_3698|M.abscessus_ATCC_1997 VVHPQLTVSQALHYAAELRLPPDTTKEDREQVVSRVLEELEMTKHAETRV
MLBr_00669|M.leprae_Br4923 RLLQQKTVYENVAFALEVIG---RRSDAINQVVPDVLETVGLSGKANRLP
: * ** . : :* *: . :** *** : :: : :
MSMEG_1642|M.smegmatis_MC2_155 DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLAD
TH_4260|M.thermoresistible__bu DKLSGGQRKRASVAMELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLAD
Mflv_4882|M.gilvum_PYR-GCK DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLAD
Mvan_1546|M.vanbaalenii_PYR-1 DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLAD
Mb1776|M.bovis_AF2122/97 DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLAD
Rv1747|M.tuberculosis_H37Rv DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLAD
MUL_3149|M.ulcerans_Agy99 DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDRQVMTMLRQLAD
MMAR_1174|M.marinum_M DKLSGGQRKRASVALELLTGPSLLILDEPTTGLDPALDRQVMLMLRQLAD
MAV_3146|M.avium_104 DKLSGGQRKRASVALELLTGPSLLILDEPTSGLDPALDLQVMTMLRQLAD
MAB_3698|M.abscessus_ATCC_1997 DKLSGGQRKRASVAMELLTGPSLLILDEPTSGLDPALDRQVMSLMRQLAD
MLBr_00669|M.leprae_Br4923 DELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERINR
*:****:::*.::* ::. * :*: **** .*** . ::* ::.::
MSMEG_1642|M.smegmatis_MC2_155 AGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFFGPPSQIGPELGTTNWAD
TH_4260|M.thermoresistible__bu AGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGPTMGTTNWAD
Mflv_4882|M.gilvum_PYR-GCK AGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGSAMGTTNWAD
Mvan_1546|M.vanbaalenii_PYR-1 AGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFYGPPSQIGPSMGTTNWAD
Mb1776|M.bovis_AF2122/97 AGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPTQIGPVMGTTNWAD
Rv1747|M.tuberculosis_H37Rv AGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPTQIGPVMGTTNWAD
MUL_3149|M.ulcerans_Agy99 AGRVVLVVTHSLTYLDVCDQVLLLAPGGKTAFCGPPSQIGPAMGTTNWAD
MMAR_1174|M.marinum_M AGRVVLIVTHSVAYLDVCDQLLLVAPGGKTAFLGPPSEISQAMGTANWAD
MAV_3146|M.avium_104 AGRVVLVVTHSLSYLDVCDQVLLMAPGGKTAYCGAPDRIGPAMGTTNWAK
MAB_3698|M.abscessus_ATCC_1997 AGRVVVVVTHSLTYLSYCDQVLLLAPGGKTAYCGPPRSIGQEMGTTDWAD
MLBr_00669|M.leprae_Br4923 TGTTVLMATHDHHIVDSMRQRVVELSLG-------------RLVRDEWCG
:* .*::.**. :. * :: . * : :*.
MSMEG_1642|M.smegmatis_MC2_155 IFSTVADDPAEANRRYLARTPEA--PPASASSEAPADLG-APAKTSLRRQ
TH_4260|M.thermoresistible__bu IFSTVAGDPDAANRRFLERQGPP--RELPAESAKPTDLG-SPTRTSLRRQ
Mflv_4882|M.gilvum_PYR-GCK IFSSVAGDPEAAAARYLSQNGPP--PPQPP-ALTPSELG-NPTRTSLRRQ
Mvan_1546|M.vanbaalenii_PYR-1 IFSAVAGDPDAAAQRYLAQNGPP--PPQPP-ALTPSELG-NPTRTSLRRQ
Mb1776|M.bovis_AF2122/97 IFSTVADDPDAAKARYLARTGPT--PPPPP-VEQPAELG-DPAHTSLFRQ
Rv1747|M.tuberculosis_H37Rv IFSTVADDPDAAKARYLARTGPT--PPPPP-VEQPAELG-DPAHTSLFRQ
MUL_3149|M.ulcerans_Agy99 IFSTVADDPEGAQARYLAQTGPP--PPAPP-VERPGELGGDPSHTSLFRQ
MMAR_1174|M.marinum_M IFTNVGADPDEANRRFLEQKQEQ--PAFPSEASPAVDLG-EPVHTDVLHQ
MAV_3146|M.avium_104 IFTQVGADPEEANRRFREQREAQQRSHKSDRADEPEDLG-EPVHTSLRHQ
MAB_3698|M.abscessus_ATCC_1997 IFANAAADPDGVHQAYLSRHPAAN--KPATAASPPGDLG-KPPKTSLLRQ
MLBr_00669|M.leprae_Br4923 IYGMDR--------------------------------------------
*:
MSMEG_1642|M.smegmatis_MC2_155 FSTIGRRQLRLIISDRGYFIFLALLPFIMGTLSLSVPGTVGFGVPN---P
TH_4260|M.thermoresistible__bu FSTIARRQIRLIVSDRGYFVFLALLPFIMGVLSLSVPGDVGFGVPN---P
Mflv_4882|M.gilvum_PYR-GCK LSTIARRQIRLVISDRGYFAFLMLLPFIMGILSLSVPGDVGFGVPKTALE
Mvan_1546|M.vanbaalenii_PYR-1 LSTIARRQIRLVISDRGYFAFLMLLPFIMGVLSLSVPGDVGFGVPKTALE
Mb1776|M.bovis_AF2122/97 FSTIARRQLRLIVSDRGYFVFLALLPFIMGALSMSVPGDVGFGFPN---P
Rv1747|M.tuberculosis_H37Rv FSTIARRQLRLIVSDRGYFVFLALLPFIMGALSMSVPGDVGFGFPN---P
MUL_3149|M.ulcerans_Agy99 FSTIARRQMRLIISDRGYFIFLAILPFVMGSLSMSVPGDVGFGMPN---P
MMAR_1174|M.marinum_M FSTVARRQIRLVISDRGYTVFLALLPFLIGTLTLTVRGKTGYGMSD---P
MAV_3146|M.avium_104 ISTIARRQVRLVVADRAYFVFLALLPFILGSLSLTVPGHNGFRVPA---P
MAB_3698|M.abscessus_ATCC_1997 VSTVARRQGRLILADRGYFIFLALLPFVLAGLVLSVPGSAGFNVALP---
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 MGDAPNEPGQILVLLNVGAIFMGTALTIRDLIGERAIFRREQAVGLSTTA
TH_4260|M.thermoresistible__bu MGDAPNEPGQILVLLNVGAIFMGTALTIRDLIGERAIFRREQAVGLSTTA
Mflv_4882|M.gilvum_PYR-GCK GGEAPNEPGQILVMLNVGAIFMGTALTIRALIGERAIFRREQAVGLSTSA
Mvan_1546|M.vanbaalenii_PYR-1 GGEAPNEPGQILVMLNVGAIFMGTALTIRALISERAIFRREQAVGLSTSA
Mb1776|M.bovis_AF2122/97 MGDAPNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLSTTA
Rv1747|M.tuberculosis_H37Rv MGDAPNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLSTTA
MUL_3149|M.ulcerans_Agy99 MGNAPNEPGQILVLLNVGAVFMGTALTIRDLIGERAIFRREQAVGLSTTA
MMAR_1174|M.marinum_M MGNNPAQPDQILVMLNVGAVFMGTALTIRDLIGERPIFRREQAVGLSTAA
MAV_3146|M.avium_104 AAGTPDEAAQILALLLPAAAFMGTALTIRDLVGERAIFQREQAVGLSTTA
MAB_3698|M.abscessus_ATCC_1997 --DNPNEPGLLLALTNLGASFLGLALTIRDLIGERVIFRREQAVGLSATA
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 YLLAKVCVYSVFAVVQSAIVTVITISGKGWG--EGAVERGVVIPN----R
TH_4260|M.thermoresistible__bu YLLAKVCVYSVFAIVQSAIVTAITIAGKGWDVDKGAVDHGSVFGN----P
Mflv_4882|M.gilvum_PYR-GCK YLLAKVVVFTAFAIVQAAIVTTIAIIGKGWG--PGAVDSGAVVGD----R
Mvan_1546|M.vanbaalenii_PYR-1 YLLAKVVVFTAFAIVQAAIVTSIAIAGKGWG--PGAVERGAVIGN----R
Mb1776|M.bovis_AF2122/97 YLIAKVCVYTVLAVVQSAIVTVIVLVGKGGP-----TQGAVALSK----P
Rv1747|M.tuberculosis_H37Rv YLIAKVCVYTVLAVVQSAIVTVIVLVGKGGP-----TQGAVALSK----P
MUL_3149|M.ulcerans_Agy99 YLLAKICVYTVFAVVQSAIVTIIVLIGKGGP-----TQGAVALGK----P
MMAR_1174|M.marinum_M YMGAKIVVFCAFAVTQAAIATTISVVGWGKP-----LSDAVLLGD----V
MAV_3146|M.avium_104 YLLAKTAVFCGFAVLQSAIVTAIVVIGKGAP-----TRGAVLFGHSTAGA
MAB_3698|M.abscessus_ATCC_1997 YLLAKIITYCGAAIVQSAIMTAVIVIVRGGP-----TQGAVLLGS----A
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 SLELFLSMAATTVTAAMVGLALSALAKTSEQIMPLLVVAIMSQLVFSGGM
TH_4260|M.thermoresistible__bu KLELFFGMAATTVAAAMLGLLLSALAKTSEQIMPLLVVAIMSQLVFSGGM
Mflv_4882|M.gilvum_PYR-GCK SIELFLDIAVTCVASAMVGLALSALAKSAEQIMPLLVVAIMSQLVFSGGM
Mvan_1546|M.vanbaalenii_PYR-1 SVELFTDIAVTCVAAAMVGLALSALAKSAEQIMPLLVVAIMSQLVFSGGM
Mb1776|M.bovis_AF2122/97 DLELFVDVAVTCVASAMLGLALSAIAKSNEQIMPLLVVAVMSQLVFSGGM
Rv1747|M.tuberculosis_H37Rv DLELFVDVAVTCVASAMLGLALSAIAKSNEQIMPLLVVAVMSQLVFSGGM
MUL_3149|M.ulcerans_Agy99 GLELFADVAATCVASAMLGLALSAVAKSNEQIMPLLVVAVMSQLVFSGGM
MMAR_1174|M.marinum_M RFELFVTVAATCVASALLGMALSSLAQSQDQIMPMLVVSIMSQLVFSGGM
MAV_3146|M.avium_104 VAELFATVAATCVASAVLGLAISSLVRSSEQIMPLFVVTVMAQLVLCGGM
MAB_3698|M.abscessus_ATCC_1997 NFELWVAVAATACAATVLGLVLSSLATSAEQVMPMLVIATMTQIVFSGGL
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 IPVTGRIGLDQLSWITPARWGFAASASTVDLIRLVPGPLTPKDSHWEHTS
TH_4260|M.thermoresistible__bu IPVTDRIGLDQLSWFTPARWGFAASASTADLITLVPGPLTPKDRHWEHDS
Mflv_4882|M.gilvum_PYR-GCK IPVTDRLVLDQMSWFTPARWGFAASASTVDLIRLVPGPLTPQDRHWEHTP
Mvan_1546|M.vanbaalenii_PYR-1 IPVTDRIVLDQMSWFTPARWGFAASASTVDLIRLVPGPLTPQDRHWQHTT
Mb1776|M.bovis_AF2122/97 IPVTGRVPLDQMSWVTPARWGFAASAATVDLIKLVPGPLTPKDSHWHHTA
Rv1747|M.tuberculosis_H37Rv IPVTGRVPLDQMSWVTPARWGFAASAATVDLIKLVPGPLTPKDSHWHHTA
MUL_3149|M.ulcerans_Agy99 IPVTDRLGLDQMSWATPARWGFATSASTVDLIKLVPGPLTPKDSHWHHTA
MMAR_1174|M.marinum_M IWVTNRFLLDQLSWLTPARWGFAASASTIDTHRLVPGPTDPKDQHWDHKA
MAV_3146|M.avium_104 VPVTGRLGLNQLSWAMPARWGYAAASATVDLRHLVPDSLLSQDRFWLHTP
MAB_3698|M.abscessus_ATCC_1997 IPVTGRAGLEQLSFLFPSRWGFAATAATCDLRKLVP--VGPQDELWNHDA
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MSMEG_1642|M.smegmatis_MC2_155 GTWLFDMAMLGVLSVFYVTFVRWKIRLKAG-
TH_4260|M.thermoresistible__bu GTWLFDMAMLAVLSVFYLALVRWKIRLPRA-
Mflv_4882|M.gilvum_PYR-GCK GTWLFDMGMLALISVGYLALVRWRIRLKSAG
Mvan_1546|M.vanbaalenii_PYR-1 GAWLFDMGMLLAISVGYLAFVRWRIRLKSA-
Mb1776|M.bovis_AF2122/97 SAWWFDMAMLVALSVIYVGFVRWKIRLKAC-
Rv1747|M.tuberculosis_H37Rv SAWWFDMAMLVALSVIYVGFVRWKIRLKAC-
MUL_3149|M.ulcerans_Agy99 SAWWFDMAMLAFISLCYVSFVRWKIRLKGG-
MMAR_1174|M.marinum_M SAWLFDMAMLAVLSIAYTAIVWWKIRLKRR-
MAV_3146|M.avium_104 GTWLLDMGMLAALALCYAVFVRWKIRLRR--
MAB_3698|M.abscessus_ATCC_1997 GTWLFDMGILGLLSLVMAGFVRWRIRLKG--
MLBr_00669|M.leprae_Br4923 -------------------------------