For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSAEPDLDVLRGRLAGAGITALTPLSGGASGLTFAGIRDGRPVVIKVAPPGVAPVGHRDVLRQARIIKAL APTDVPVPRVCWEDAGRPPYTPPLYVMSRVEGDCGEPLFDGWPAVPGLADRYRNACRTMAALHRIPPTEL GLGGEPVIDPVAEVHRWSDTLGTVDPALAPGWPKVRDALLDCAPRAMPPAVVHGDFRLGNLLADGPRINA VIDWEIWSIGDPRIDVGWFLINCDPDTYRRVTPSDGAVPSTAELADLYLHELGCDAGDLDWFSALACFKS AATWSLIVKHNRRRSSPRAELESMTTTLPRLLDRAGALLARRR*
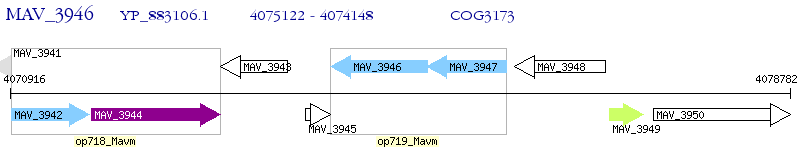
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3946 | - | - | 100% (324) | phosphotransferase enzyme family protein, putative |
| M. avium 104 | MAV_0312 | - | 3e-22 | 27.64% (322) | phosphotransferase enzyme family protein |
| M. avium 104 | MAV_5273 | - | 4e-22 | 29.01% (293) | phosphotransferase enzyme family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3787c | fadE36 | 8e-20 | 28.35% (321) | acyl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4318 | - | 2e-20 | 27.74% (292) | aminoglycoside phosphotransferase |
| M. tuberculosis H37Rv | Rv3761c | fadE36 | 8e-20 | 28.35% (321) | acyl-CoA dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4910c | - | 4e-24 | 28.77% (292) | putative aminoglycoside phosphotransferase |
| M. marinum M | MMAR_5325 | - | 1e-21 | 29.34% (317) | hypothetical protein MMAR_5325 |
| M. smegmatis MC2 155 | MSMEG_6337 | - | 1e-20 | 29.43% (299) | phosphotransferase enzyme family protein |
| M. thermoresistible (build 8) | TH_2427 | - | 5e-53 | 38.69% (305) | PUTATIVE putative phosphotransferase enzyme family protein, |
| M. ulcerans Agy99 | MUL_4404 | - | 1e-21 | 30.10% (289) | hypothetical protein MUL_4404 |
| M. vanbaalenii PYR-1 | Mvan_2028 | - | 1e-22 | 27.27% (308) | aminoglycoside phosphotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3946|M.avium_104 ------MSSAEPDLDVLRGRLAG-------AGITALTPLSGGASGLTFAG
TH_2427|M.thermoresistible__bu ----MSVNTANAASAALVSRITERFARDLDLRVSEVTPIVGGHSGLTYRV
Mb3787c|M.bovis_AF2122/97 --MTSVDRLDGLDLGALDRYLRSL--GIGRDGELRGELISGGRSNLTFRV
Rv3761c|M.tuberculosis_H37Rv --MTSVDRLDGLDLGALDRYLRSL--GIGRDGELRGELISGGRSNLTFRV
MSMEG_6337|M.smegmatis_MC2_155 --MTS---LEGLDLDALDRHLRAE--GIARAGDLRAELIAGGRSNLTFLV
Mflv_4318|M.gilvum_PYR-GCK --MPVDVTLTDDAVAALQEWVRRT--GLG-SVVSDVEPLTGGSQNIVVRV
Mvan_2028|M.vanbaalenii_PYR-1 --MPVDVTLTEAETDALRDWVRRT--GIG-STVTGVEPLTGGSQNIVVRL
MMAR_5325|M.marinum_M ---MHPVDP-----DAVAEWMSGQ--GLGEGPLYDVCAVTGGTQNVMLRF
MUL_4404|M.ulcerans_Agy99 --------------------MSGQ--GLGEGPLYDVCAVTGGTQNVMLRF
MAB_4910c|M.abscessus_ATCC_199 MAAPHHIDPKSVDFDVVSHWMDEQ--GLPGGQLTDISAITGGTQNIMVRF
: : ** ..:
MAV_3946|M.avium_104 IRDGRPVVIKVAPPGVAPVGHRDVLRQARIIKALAPTDVPVPRVCWEDAG
TH_2427|M.thermoresistible__bu DTDGPAYVVKAVPEGQRPIGRHDMMRQARILRALADTPVPVPAVAVVDET
Mb3787c|M.bovis_AF2122/97 YDDASSWLVRRPPLHGLTPSAHDMAREYRVVAALGDTPVPVARTISLCQD
Rv3761c|M.tuberculosis_H37Rv YDDASSWLVRRPPLHGLTPSAHDMAREYRVVAALGDTPVPVARTISLCQD
MSMEG_6337|M.smegmatis_MC2_155 FDDASKWVLRRPPLHGLTPSAHDMAREYRVVAALAGTPVPVARAVTMRND
Mflv_4318|M.gilvum_PYR-GCK RLDGSDVVLRRPPQHPRPTSNDTMRREIAVLQTLRGTDAPHPEFVAGCED
Mvan_2028|M.vanbaalenii_PYR-1 QVDGRPMVLRRPPPHPRPTSDNTMRREIAVLQTLKGTAVPHPELIAGCED
MMAR_5325|M.marinum_M TRSGRTYVLRRGPRHLRPRSNSVILRETKVLAALAGTDVPHPRLIAACPD
MUL_4404|M.ulcerans_Agy99 TRSGRTYVLRRGPRHLRPRSNSVILRETKVLAALAGTDVPHPRLIAACPD
MAB_4910c|M.abscessus_ATCC_199 SRGGREYVLRRPPKHLREASNNVIRREARLLGALRGQGVPAPELIAACTD
.. ::: * . : *: :: :* .* .
MAV_3946|M.avium_104 RPPYT-PPLYVMSRVEGDCGEPLFDGWPAVPG-LADRYRNACR-TMAALH
TH_2427|M.thermoresistible__bu AP-----AWFAMELVPGDSLEPVLDDPPVPPKRAARRMRAAAG-LLKTLH
Mb3787c|M.bovis_AF2122/97 DSVLG-APFQVVEFVAGQVVRRRAELEALGSRSVIEGCVDALIRVLVDLH
Rv3761c|M.tuberculosis_H37Rv DSVLG-APFQVVEFVAGQVVRRRAELEALGSRSVIEGCVDALIRVLVDLH
MSMEG_6337|M.smegmatis_MC2_155 DSVLG-APFQMVDHVDGRVVRTAAELAALGDQTVIDNCIDALIKALSDLH
Mflv_4318|M.gilvum_PYR-GCK LDVLG-VVFYLMQAVDGFNPGTEIDEAYVRDAGMRHRVGLSYAASLARLG
Mvan_2028|M.vanbaalenii_PYR-1 LDVLG-VVFYLMQAIDGFNPGTEIDEAYIRDAGMRHRAGLSYAASLAELG
MMAR_5325|M.marinum_M PTVLGDAVFYLMEPVEGFNAGEGLPPLHAGDASIRFQMGLSMADALAKLG
MUL_4404|M.ulcerans_Agy99 PTVLDDAVFYLMEPVEGFNAGEGLPPLHAGDASIRFQMGLSMADALAKLG
MAB_4910c|M.abscessus_ATCC_199 ETVLGGAVFYLMEPVDGFNASVGLPELHAGNAEIRHQMGLEAVSGIAALG
:. : * : *
MAV_3946|M.avium_104 RIPPTELGLG--GEPVIDPVAEVHRWSDTLGTVD------PALAPGWPKV
TH_2427|M.thermoresistible__bu AVPVDSIPTD--AAPLS-PEDELARWARTMGAVP------PELVPRAEEL
Mb3787c|M.bovis_AF2122/97 SIDPKAVGLSDFGKPDGYLERQVRRWGSQWELVRLPDDHRDADIS---RL
Rv3761c|M.tuberculosis_H37Rv SIDPKAVGLSDFGKPDGYLERQVRRWGSQWELVRLPDDHRDADIS---RL
MSMEG_6337|M.smegmatis_MC2_155 AVDPYAVGLGDFGKPDGYLERQVRRWGSQWEHVRLPDDARDDDVR---RL
Mflv_4318|M.gilvum_PYR-GCK AVPWEGSPLAALKRPGSFLARQVPQFLRLLESYR-HENYAPESFPSVHVL
Mvan_2028|M.vanbaalenii_PYR-1 KVPWEGSPLAALKRPGSFLARQVPQFMRLLESYR-HERYAPESFPSVHLL
MMAR_5325|M.marinum_M AVDHAAVGLADFGKPAGFLERQVPRWLAELESYQQYDGYPGPDIPGIGEV
MUL_4404|M.ulcerans_Agy99 AVDHAAVGLADFGKPAGFLERQVPRWLAELESYQQYDGYPGPDIPGIGEV
MAB_4910c|M.abscessus_ATCC_199 ALDYQELGLDGYGNPDGFLERQVPRWLKELDSYAKHDGYPGPDIPGLQSV
: * :: :: :
MAV_3946|M.avium_104 RDALLDCAPRAMPPAVVHGDFRLGNLLAD---GPRINAVIDWEIWSIGDP
TH_2427|M.thermoresistible__bu QRRLAEDVPAAMPVTLVHGDFRLGNIIAD---SDDLAALIDWEIWSLGDP
Mb3787c|M.bovis_AF2122/97 HLALQQAIPQQSRTSIVHGDYRIDNTILDTDDPCHVRAVVDWELSTLGDP
Rv3761c|M.tuberculosis_H37Rv HLALQQAIPQQSRTSIVHGDYRIDNTILDTDDPCHVRAVVDWELSTLGDP
MSMEG_6337|M.smegmatis_MC2_155 HAALADAVPPQSGTSIVHGDYRIDNTVLDAEDATVVRAVLDWEMSTLGDP
Mflv_4318|M.gilvum_PYR-GCK AEWLEAHRPPDSEPGIMHGDCHLNNVLLRRDVP-ELAAFIDWEMCTVGDP
Mvan_2028|M.vanbaalenii_PYR-1 AEWLESHRPEDTEPGIMHGDCHLSNVLLRRDVP-EVAAFVDWEMCTVGDP
MMAR_5325|M.marinum_M SCWLDSHRPTAWTPGILHGDYHMANVMFSRTGP-EVVAICDWEMCTIGDP
MUL_4404|M.ulcerans_Agy99 SCWLDSHRPTAWTPGILHGDYHMANVMFSRTGP-EVVAICDWEMCTIGDP
MAB_4910c|M.abscessus_ATCC_199 ANWLERHRPSQWKPGIMHGDFHLANMMFRNDGP-QLAAIVDWEMSTIGDP
* * ::*** :: * : : *. ***: ::***
MAV_3946|M.avium_104 RIDVGWFLINCD----PDTYRRVTPSDGAVPSTAELADLYLHELGCDAGD
TH_2427|M.thermoresistible__bu RVELGWFLVFAD----GTNFPGAGRVVPGLPGADELRGIYTETP---LRD
Mb3787c|M.bovis_AF2122/97 LSDAALMCVYRDPALDLIVHAQAAWTSPLLPAADELADRYSLVSGQPLGH
Rv3761c|M.tuberculosis_H37Rv LSDAALMCVYRDPALDLIVHAQAAWTSPLLPAADELADRYSLVSGQPLGH
MSMEG_6337|M.smegmatis_MC2_155 LSDAALMCVYRNPMFG-HIHTDAAWSSDLVPPADELADRYSKVSGRDLAH
Mflv_4318|M.gilvum_PYR-GCK LLDLGWLLVCWPDGPNPIDAGSALGALGGLASRAELIDAYRDAGGRPTTH
Mvan_2028|M.vanbaalenii_PYR-1 LLDLGWMLICWPDGPNPIDVGSELAALGGLATRPELVEAYLAAGGRRTSA
MMAR_5325|M.marinum_M LLDLGWLLATWRQSDGASVFGHALGAQDGLASTDDLLQRYAANTTRDLSH
MUL_4404|M.ulcerans_Agy99 LLDLGWLLATWRQSDGASVFGHALGAQDGLASTDDLLQRYAANTTRDLSH
MAB_4910c|M.abscessus_ATCC_199 LLDLGWMLATWPSETDDASLGGALVQAGGLPTPQELVAHYAERTDRDLGA
: . : :. :* *
MAV_3946|M.avium_104 LDWFSALACFKSAATWSLIVKHNRRRSSPRAEL---ESMTTTLPRLLDRA
TH_2427|M.thermoresistible__bu VDWFDALGRFKMAAIMGHNLRRHREGRHHDPEQ---EKLPATIRSLIDNG
Mb3787c|M.bovis_AF2122/97 WEFYMALAYFKLAIIAAGIDYRRRMSEQAEGKDTAAESVPDVVAPLIARG
Rv3761c|M.tuberculosis_H37Rv WEFYMALAYFKLAIIAAGIDYRRRMSEQAEGKDTAAESVPDVVAPLIARG
MSMEG_6337|M.smegmatis_MC2_155 WDFYMALAYFKLAIIAAGIDYRRR--EGADG----PEQVGEAVAPMIAAG
Mflv_4318|M.gilvum_PYR-GCK LDWYIAMACFKLAIVIEGTWSRYLAGRASAEAG---ERLHASAENLIALG
Mvan_2028|M.vanbaalenii_PYR-1 LDWYVAMACFKLAIVIEGTWSRYLAGQASAEAG---ERLHASAENLIELG
MMAR_5325|M.marinum_M ITWYTVLACFKLGIVIEGTLARACAGQAEKEVG---DQLHAATVHLFERA
MUL_4404|M.ulcerans_Agy99 ITWYTVLACFKLGIIVEGTLARACAGQAEKEVG---DQLHAATVHLFERA
MAB_4910c|M.abscessus_ATCC_199 MDWYTVLACFKLGIILEGTHARACAGKAPVAVG---DYLHGLTLQLFKRA
:: .:. ** . : : : :: .
MAV_3946|M.avium_104 GALLARRR---
TH_2427|M.thermoresistible__bu LRLLG------
Mb3787c|M.bovis_AF2122/97 LAEIAKKSG--
Rv3761c|M.tuberculosis_H37Rv LAEIAKKSG--
MSMEG_6337|M.smegmatis_MC2_155 LAALG------
Mflv_4318|M.gilvum_PYR-GCK TRVSTGDNPFV
Mvan_2028|M.vanbaalenii_PYR-1 TRVSKGDNPFA
MMAR_5325|M.marinum_M LGLIGA-----
MUL_4404|M.ulcerans_Agy99 LGLIGA-----
MAB_4910c|M.abscessus_ATCC_199 VAITG------