For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAGLPGIRGWQEDLYRDLHRHPELSHQEHRTAAVVAERLRGFGYDVHEGVGGTGVVGVLRNGTGPAVLLR ADMDALPVREDTGLPYASAVTATDPRGDERPVMHACGHDVHVTCLLGAASLLAGAREHWRGSLVAVFQPA EELGDGARRMVDDGLARLVGHVDVALAQHVLPFPAGQIVTRSGPFMATADSMRVTVYGRGGHGSMPQTTV DPVVLAAMIVIRLQTVVSREVAPTETAVLTIGRIESGAKSNVIGDHAVLELNLRTFSGRTRDRMLDAVRR IVTAECQASGSPRDPEFEMMNHFPATDNDPAVTARVRAVFAEHFGDRATEIPAQTGSEDFSNIATGLDAP YSYWGFGGADPGAYRRAEAAGRLAEDIPVNHSGRFAPVIQPTLDTGTEALTVAALAWLAPSAS*
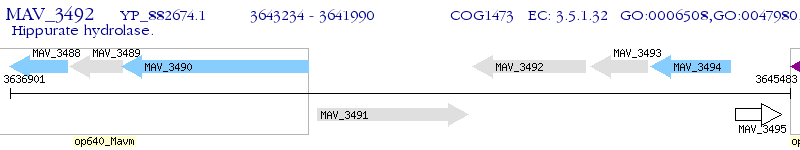
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3492 | - | - | 100% (414) | N-alpha-acyl-glutamine aminoacylase |
| M. avium 104 | MAV_4281 | - | 9e-39 | 35.37% (328) | amidohydrolase |
| M. avium 104 | MAV_4282 | - | 8e-11 | 23.04% (408) | amidohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3333c | amiA1 | 4e-39 | 38.27% (277) | N-acyl-L-amino acid amidohydrolase |
| M. gilvum PYR-GCK | Mflv_1971 | - | 1e-165 | 69.17% (412) | amidohydrolase |
| M. tuberculosis H37Rv | Rv3305c | amiA1 | 4e-39 | 38.27% (277) | N-acyl-L-amino acid amidohydrolase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4738c | - | 1e-113 | 51.00% (402) | putative amidohydrolase |
| M. marinum M | MMAR_1221 | amiA1 | 1e-36 | 37.18% (277) | N-acyl-L-amino acid amidohydrolase AmiA1 |
| M. smegmatis MC2 155 | MSMEG_3989 | - | 3e-57 | 39.72% (355) | peptidase M20D, amidohydrolase |
| M. thermoresistible (build 8) | TH_1760 | - | 6e-36 | 38.64% (264) | POSSIBLE N-ACYL-L-AMINO ACID AMIDOHYDROLASE AMIA1 |
| M. ulcerans Agy99 | MUL_2671 | amiA1 | 2e-36 | 36.82% (277) | N-acyl-L-amino acid amidohydrolase AmiA1 |
| M. vanbaalenii PYR-1 | Mvan_4743 | - | 1e-166 | 69.42% (412) | amidohydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1221|M.marinum_M ---MSLVDTAES-WLAAHYDDLVGWRRHIHRYPELGRQEFATTQFVAERL
MUL_2671|M.ulcerans_Agy99 ---MSLVDTAES-WLAAHYDDLVGWRRHIHRYPELGRQEFATTQFVAERL
Mb3333c|M.bovis_AF2122/97 ---MSLADAAES-WLAAHHDDLVGWRRHIHRYPELGRQEYATTQFVAERL
Rv3305c|M.tuberculosis_H37Rv ---MSLADAAES-WLAAHHDDLVGWRRHIHRYPELGRQEYATTQFVAERL
TH_1760|M.thermoresistible__bu ---MSVLRHAVARWLSRHQDDLVAWRRHIHAHPELGRQEFATTQFVAERL
Mflv_1971|M.gilvum_PYR-GCK MAPSSTAATAVLDGLPGIRAWQEDFYRDLHRHPELSHQEHRTSAKAAEAL
Mvan_4743|M.vanbaalenii_PYR-1 MAPSS-QATTVLDGLPGIRSWQEDFYRDLHRHPELSHQEHRTAGKVVDTL
MAV_3492|M.avium_104 ----------MLAGLPGIRGWQEDLYRDLHRHPELSHQEHRTAAVVAERL
MAB_4738c|M.abscessus_ATCC_199 ------MSITLPHNWADDLGDLADFYRDLHQNPELSFQEHRTAGKIEEAI
MSMEG_3989|M.smegmatis_MC2_155 --------MSLVTQAREMADDLVHLRRALHQEPEVGLNLPRTQERVLAAL
* :* **:. : * :
MMAR_1221|M.marinum_M ADAGLNPKILPGGTGLTCDFGPEHQPRIALRADMDALPMAERTGAPYAST
MUL_2671|M.ulcerans_Agy99 ADAGLNPKILPGGTGLTCDFGPEHQPRIALRADMDALPMAERTGAPYAST
Mb3333c|M.bovis_AF2122/97 ADAGLNPKVLPGGTGLTCDFGPQHQPRIALRADMDALPMAERTGAPYAST
Rv3305c|M.tuberculosis_H37Rv ADAGLNPKVLPGGTGLTCDFGPQHQPRIALRADMDALPMAERTGAPYAST
TH_1760|M.thermoresistible__bu ADAGLNPKVLPGGTGLICDLGPDHTPRVALRADMDALPMPERTGAPYAST
Mflv_1971|M.gilvum_PYR-GCK --AGYRVHTGIGGTGVIGILDNGPGPTVLLRADMDALPVLEHTGLPYASS
Mvan_4743|M.vanbaalenii_PYR-1 TAAGYRVHDGIGGTGVIGILENGPGPTVLLRADMDALPVLEDTGLPYAST
MAV_3492|M.avium_104 RGFGYDVHEGVGGTGVVGVLRNGTGPAVLLRADMDALPVREDTGLPYASA
MAB_4738c|M.abscessus_ATCC_199 APLGLEVTAGIGGTGVVAVLRNGSGPVIWLRADFDALPVQEQTGLPYAST
MSMEG_3989|M.smegmatis_MC2_155 EGLPLEITTGTNCTSVTAVLRGHGDGVVLLRADMDALPVQEHTGLDYTSR
. *.: : : ****:****: * ** *:*
MMAR_1221|M.marinum_M MPN---------IAHACGHDGHTAILLGTALALATVPELPVGVRLIFQAA
MUL_2671|M.ulcerans_Agy99 MPN---------IAHACGHDGHTAILLGTALALATVPELPVGVRLIFEAA
Mb3333c|M.bovis_AF2122/97 MPN---------VAHACGHDAHTAILLGAALALASVPELPVGVRLIFQAA
Rv3305c|M.tuberculosis_H37Rv MPN---------VAHACGHDAHTAILLGAALALASVPELPVGVRLIFQAA
TH_1760|M.thermoresistible__bu VPN---------VAHACGHDAHTAMLLGAALALNTVPELPIGVRLIFQAA
Mflv_1971|M.gilvum_PYR-GCK ERATDATGHEVPVMHACGHDAHVTALLGAARLMADGRSNWGGTLVALFQP
Mvan_4743|M.vanbaalenii_PYR-1 ERATDPTGHEVPVTHACGHDLHVTALLGATRLLADGRGSWGGTLVGLFQP
MAV_3492|M.avium_104 VTATDPRGDERPVMHACGHDVHVTCLLGAASLLAGAREHWRGSLVAVFQP
MAB_4738c|M.abscessus_ATCC_199 VRATNPDGKDVPVMHACGHDMHVAALVGALRLLDQLKATWSGTVVAVFQP
MSMEG_3989|M.smegmatis_MC2_155 IDG---------AMHGCGHDLHTAMLVGAAHLLAAQREKLHGDVVFMFQP
*.**** *.: *:*: : * : . .
MMAR_1221|M.marinum_M EELMPGGAIDAIAAGALTGVSR----IFALHCDPR-LEVGRVAVRQGPIT
MUL_2671|M.ulcerans_Agy99 EELMPGGAIDAIAAGALTGVSR----IFALHCDPR-LEVGRVAVRQGPIT
Mb3333c|M.bovis_AF2122/97 EELMPGGAIDAIAAGALAGVSR----IFALHCDPR-LEVGKVAVRQGPIT
Rv3305c|M.tuberculosis_H37Rv EELMPGGAIDAIAAGALAGVSR----IFALHCDPR-LEVGKVAVRQGPIT
TH_1760|M.thermoresistible__bu EELMPGGALDAIAAGVLNGVSR----IFALHCDPR-LQVGRVATRPGPIT
Mflv_1971|M.gilvum_PYR-GCK AEETGEGARGMVDDGLAKLLPH-VDVCLGQHVMP--FPAGTIGVHAGPFM
Mvan_4743|M.vanbaalenii_PYR-1 AEETGDGARGMVADGLANLLSH-VDVCLGQHVMP--FPAGTIGMHAGPFM
MAV_3492|M.avium_104 AEELGDGARRMVDDGLARLVGH-VDVALAQHVLP--FPAGQIVTRSGPFM
MAB_4738c|M.abscessus_ATCC_199 AEEVGGGANAMIADGILDRFPR-PDVVLGQHVGP--LPAGMIAYKTGTAL
MSMEG_3989|M.smegmatis_MC2_155 GEEGWEGARTMIDEGVLDAAGRRPDAAYALHVFSTLGPAGTFFSRPGVAL
* ** : * : . * . .* . : *
MMAR_1221|M.marinum_M SAADHIEITLYSPGGHTSRPHLTADLVYGLGTLITGLPGVLSRRVDPRNS
MUL_2671|M.ulcerans_Agy99 SAADHIEITLYSPGGHTSRPHLTADLVYGLGTLITGLPGVLSRRVDPRNS
Mb3333c|M.bovis_AF2122/97 SAADSIEITLYSPGGHTSRPHLTADLVYGLGTLVTGLPGVLSRRIDPRNS
Rv3305c|M.tuberculosis_H37Rv SAADSIEITLYSPGGHTSRPHLTADLVYGLGTLVTGLPGVLSRRIDPRNS
TH_1760|M.thermoresistible__bu SAADHVEVTLQSKGGHTSRPHLTGDLVYALGTLITGVPGVLSRRIDPRNG
Mflv_1971|M.gilvum_PYR-GCK ASADSMRITVYGRGSHGSMPQASVDPVVLAAMIVVRLQTVVSREVSPAEP
Mvan_4743|M.vanbaalenii_PYR-1 ASADSMRVTVYGRGSHGSMPQASVDPVVLAAMIVVRLQTVVSREVSPVEP
MAV_3492|M.avium_104 ATADSMRVTVYGRGGHGSMPQTTVDPVVLAAMIVIRLQTVVSREVAPTET
MAB_4738c|M.abscessus_ATCC_199 AASDSLKLQLFGRGGHGSQPESTIDPVVMASSVVQRLQTIVSRELSPFEP
MSMEG_3989|M.smegmatis_MC2_155 AASATLRVTVRGEGGHGSTPHLAKDPVPVLAEMVTALQTAVTRQFDVFDP
::: :.: : . *.* * *. : * * . :: : ::*.. :
MMAR_1221|M.marinum_M TVLVWGAVNAGVAANAIPQTGVLAGTVRTASRQTWIDLEEIIRESITALL
MUL_2671|M.ulcerans_Agy99 TVLVWGAVNAGVAANAIPQTGVLAGTVRTASRQTWIDLEEIIRESITALL
Mb3333c|M.bovis_AF2122/97 TVLVWGAVNAGMAANAIPQTGVLSGTVRTASRQTWVDLEELVRQAISALL
Rv3305c|M.tuberculosis_H37Rv TVLVWGAVNAGMAANAIPQTGVLSGTVRTASRQTWVDLEELVRQAISALL
TH_1760|M.thermoresistible__bu TVMVWGAVNAGVAANAIPQTGTLAGTIRTASRSTWLTLEAIVSETVSSLL
Mflv_1971|M.gilvum_PYR-GCK AVLTVGSITAGSKSNVIPDRAVLQLNIRTFNDATRTAVLDAVRRIVTAEC
Mvan_4743|M.vanbaalenii_PYR-1 VVLTVGSITSGSKSNVIPDRAVLQLNIRTFNDDTRTAVLEAVRRIVTAEC
MAV_3492|M.avium_104 AVLTIGRIESGAKSNVIGDHAVLELNLRTFSGRTRDRMLDAVRRIVTAEC
MAB_4738c|M.abscessus_ATCC_199 AVVTVGYVHAGAKDNIIPDDAELGINVRTFNEEVRKKTLASITRIAQAES
MSMEG_3989|M.smegmatis_MC2_155 VVLTIGVLRAGTRSNIIPATATFEATIRTFSVESSRRIRDIALRLVSGIA
.*:. * : :* * * . : .:** . . .
MMAR_1221|M.marinum_M SP--LAIEHTLQYRRGVPPVVNEEVSTHILTHAIEAVSP--DALADTRQS
MUL_2671|M.ulcerans_Agy99 SP--LAIEHTLQYRRGVPPVVNEEVSTHILTHAIEAVSP--DALADTRQS
Mb3333c|M.bovis_AF2122/97 LP--LAIEHTLQYRRGVPPVVNEEISTRILAHAIEAIGP--GVLADTRQS
Rv3305c|M.tuberculosis_H37Rv LP--LAIEHTLQYRRGVPPVVNEEISTRILAHAIEAIGP--GVLADTRQS
TH_1760|M.thermoresistible__bu AP--LGVEHSLQYRRGVPPVVNEEQSTRILTRAIEAVGP--NVLADTRQS
Mflv_1971|M.gilvum_PYR-GCK MASRSPRDPEFELSDRFPLTDNDIPTTERVTHAFTDMFG-ERCVELP-QG
Mvan_4743|M.vanbaalenii_PYR-1 AASRSPRDPEFELSDRFPVTDNDIATTERVTHAFSEMFG-ERCVELP-QG
MAV_3492|M.avium_104 QASGSPRDPEFEMMNHFPATDNDPAVTARVRAVFAEHFG-DRATEIP-AQ
MAB_4738c|M.abscessus_ATCC_199 IASGADKEPAVTSMHTFPVTVVDDGAMQAIAAVFREHFGAERVVESPSPL
MSMEG_3989|M.smegmatis_MC2_155 DAHGVDVDADYAEGR--PPTVNDPDETALAREVIIETLGENRYAPLANPF
. : * . : .: .
MMAR_1221|M.marinum_M GGGEDFSWYLEEVPGAMGRLGVWSGDGPQLDLHQPTFDLDERALGIGVRV
MUL_2671|M.ulcerans_Agy99 GGGEDFSWYLEEVPGAMGRLGVWSGDGPQLDLHQPTFDLDERALGIGLRV
Mb3333c|M.bovis_AF2122/97 GGGEDFSWYLEEVPGAMARLGVWSGDGLQLDLHQPTFDIDERALAIGLRV
Rv3305c|M.tuberculosis_H37Rv GGGEDFSWYLEEVPGAMARLGVWSGDGLQLDLHQPTFDIDERALAIGLRV
TH_1760|M.thermoresistible__bu GGGEDFSWYLEEVPGAMARLGVWSGQGPQLDLHQPTFDLDERALPVGVRV
Mflv_1971|M.gilvum_PYR-GCK AGSEDFSDIPRALGVPYTYWGVGCVDPDVYRTAAEAGSINQDIAVNHSPR
Mvan_4743|M.vanbaalenii_PYR-1 VGSEDFSEIPRALGVPYTYWGVGCVDPDAYRSAAEAGSVNQDIAVNHSPR
MAV_3492|M.avium_104 TGSEDFSNIATGLDAPYSYWGFGGADPGAYRRAEAAGRLAEDIPVNHSGR
MAB_4738c|M.abscessus_ATCC_199 AGSEDVGIFGTTAGVPTAFWFWGGYEQQRFEEAAAAG---KPVPSNHSPE
MSMEG_3989|M.smegmatis_MC2_155 TGAEDFARVLD--EVPGCFVALGALPPGADPDKAAYNHSPQAVFDDSVLP
*.**.. . :
MMAR_1221|M.marinum_M MVNIIEQAAAF-------------------
MUL_2671|M.ulcerans_Agy99 MVNIIEQAAAF-------------------
Mb3333c|M.bovis_AF2122/97 MVNIIEQAAAH-------------------
Rv3305c|M.tuberculosis_H37Rv MVNIIEQAAAH-------------------
TH_1760|M.thermoresistible__bu FVNLVEQCAA--------------------
Mflv_1971|M.gilvum_PYR-GCK FAPVIQPTLDTATQALITAALAWLTPA---
Mvan_4743|M.vanbaalenii_PYR-1 FAPVIQPTLDTATQALVAAAMTWLAPT---
MAV_3492|M.avium_104 FAPVIQPTLDTGTEALTVAALAWLAPSAS-
MAB_4738c|M.abscessus_ATCC_199 FAPVLEPTLSTGIEALTVAALSRLTARA--
MSMEG_3989|M.smegmatis_MC2_155 DGAALYAELAHRRLNVNSGATSGATTQKGR
: