For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTEIRIESLGAISAAVGEFDRGLTVLTGETGTGKTMVVTGLHLLGGARADATRVRSGADRAIVEGRFTTT DLDEALVVRLDEMLDASGAERDEDGSVIALRSVNRDGPSRAYLGGRSVPAKSLGDFTAELLTLHGQNDQL RLMRPEEQCGALDRFAKAGPALERYTKLRDAWLSARRDLADRRNRMRELALEADRLTFALGEIDAVDPQP GEDDALVAEIMRLSELDTLREAAAAARAALSADDADGSGLSAVDTLGKARAALESTDDPKLLALAGQIGE VLTVVVDAAGELAGFLEELPVDASALEAKLARQSELRGLTRKYAADIDGVLAWARESRERLAQLDVSEEG LSALAARVEELGRELAGAAADLSTIRRKAAKRLAKEVTAELSGLAMADAQFSIDVSTDVVPAGSGSDDAA VLTLPSGERVRAGADGIDQVEFGFAAHRGMDRLPLAKSASGGELSRVMLALEVVLAASRKETVGTTMVFD EVDAGVGGRAAVQIGRRLARLARTHQVIVVTHLPQVAAYADVHLVVHGAGPRGTSVVRRVTGDERVAELA RMLAGLGESDSGRAHARELLDAAQKDEI*
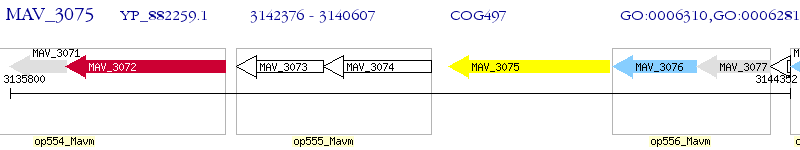
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3075 | recN | - | 100% (589) | DNA repair protein RecN |
| M. avium 104 | MAV_3777 | smc | 2e-09 | 22.82% (447) | chromosome segregation protein SMC |
| M. avium 104 | MAV_3135 | uvrA | 5e-07 | 26.30% (308) | excinuclease ABC subunit A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1722 | recN | 0.0 | 76.78% (590) | DNA repair protein recN (recombination protein N) |
| M. gilvum PYR-GCK | Mflv_3507 | - | 0.0 | 70.88% (601) | DNA repair protein RecN |
| M. tuberculosis H37Rv | Rv1696 | recN | 0.0 | 76.78% (590) | DNA repair protein recN (recombination protein N) |
| M. leprae Br4923 | MLBr_01360 | recN | 0.0 | 77.29% (590) | recombination and DNA repair |
| M. abscessus ATCC 19977 | MAB_2361 | - | 0.0 | 68.75% (592) | DNA repair protein RecN |
| M. marinum M | MMAR_2501 | recN | 0.0 | 77.38% (588) | DNA repair protein, RecN |
| M. smegmatis MC2 155 | MSMEG_3808 | uvrA | 2e-07 | 27.18% (298) | excinuclease ABC subunit A |
| M. thermoresistible (build 8) | TH_1548 | recN | 0.0 | 70.88% (594) | PROBABLE DNA REPAIR PROTEIN RECN (RECOMBINATION PROTEIN N) |
| M. ulcerans Agy99 | MUL_1685 | recN | 0.0 | 77.55% (588) | DNA repair protein, RecN |
| M. vanbaalenii PYR-1 | Mvan_3288 | - | 0.0 | 71.09% (595) | DNA repair protein RecN |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3507|M.gilvum_PYR-GCK MLAEIRIEALGAINAATAEFDG---GLTVLTGETGAGKTMVVTGLHLLGG
Mvan_3288|M.vanbaalenii_PYR-1 MLAEIRIEALGAISAATAEFDG---GLTVLTGETGAGKTMVVTGLHLLGG
TH_1548|M.thermoresistible__bu VLSEIRIESLGAISSATAEFDR---GLTVLTGETGTGKTMVVTGLHLLGG
Mb1722|M.bovis_AF2122/97 MLTELRIESLGAISVATAEFDR---GFTVLTGETGTGKTMVVTGLHLLGG
Rv1696|M.tuberculosis_H37Rv MLTELRIESLGAISVATAEFDR---GFTVLTGETGTGKTMVVTGLHLLGG
MMAR_2501|M.marinum_M MLTEIRIESLGAISAATAEFDR---GLTVLTGETGTGKTMVVTGLHLLGG
MUL_1685|M.ulcerans_Agy99 MLTEIRIESLGAISAATAEFDR---GLTVLTGETGTGKTMVVTGLHLLGG
MAV_3075|M.avium_104 MLTEIRIESLGAISAAVGEFDR---GLTVLTGETGTGKTMVVTGLHLLGG
MLBr_01360|M.leprae_Br4923 MLTEIRIESLGAISVATAEFDR---GLTVLTGETGTGKTMVVTGLHLLGG
MAB_2361|M.abscessus_ATCC_1997 MLTEIRIESLGAIPAATAEFDS---GLTVLTGETGAGKTMVVTSLHLLGG
MSMEG_3808|M.smegmatis_MC2_155 MADRLVVKGAREHNLRGVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
: .: ::. ::* .: *:** :*:**: :. . : *
Mflv_3507|M.gilvum_PYR-GCK ARADATKVRSGSDRAVVEGRFTTTEVGDAVSARVDGILDSSGAE-RDDDG
Mvan_3288|M.vanbaalenii_PYR-1 ARADATKVRSGADRAVVEGRFTTAEVGDAVAAQVDGILDSSGAE-RDDDG
TH_1548|M.thermoresistible__bu GRADPTRVRSGAQRAVVEGRFSTAELLDEAAARVDDILAASGAE-RDEDG
Mb1722|M.bovis_AF2122/97 ARADATRVRSGADRAVVEGRFTTTDLDDATVAGLQAVLDSSGAE-RDEDG
Rv1696|M.tuberculosis_H37Rv ARADATRVRSGADRAVVEGRFTTTDLDDATVAGLQAVLDSSGAE-RDEDG
MMAR_2501|M.marinum_M ARADANRVRSGADRAIVEGRFTTVDLDQSATAHLDHMLDASGAD-RDEDG
MUL_1685|M.ulcerans_Agy99 ARADANRVRSGADRAIVEGRFTTVDLDQSATAHLDHMLDASGAD-RDEDG
MAV_3075|M.avium_104 ARADATRVRSGADRAIVEGRFTTTDLDEALVVRLDEMLDASGAE-RDEDG
MLBr_01360|M.leprae_Br4923 ARADASRVRSGANRAVVEGRFTTTDLDDVVVAQLDGILNASGSE-RDEDG
MAB_2361|M.abscessus_ATCC_1997 ARADANRVRAGADRAVVEGRFLTADPLGAEPAEVTEVLDSSGAQ-RDDDG
MSMEG_3808|M.smegmatis_MC2_155 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
* : : : : : : .: : ::. : *. *
Mflv_3507|M.gilvum_PYR-GCK SVIAARSVS-----RDGPSRAYLGGRSVPAKSLSGFTNELLTLHG--QND
Mvan_3288|M.vanbaalenii_PYR-1 SVIAARSVS-----RDGPSRAYLGGRSVPAKSLSGFTNELLTLHG--QND
TH_1548|M.thermoresistible__bu TVIAARTVS-----RDGPSRAYLGGRSVPAKSLGNFTGELLTVHG--QND
Mb1722|M.bovis_AF2122/97 SVIALRSIS-----RDGPSRAYLGGRGVPAKSLSGFTNELLTLHG--QND
Rv1696|M.tuberculosis_H37Rv SVIALRSIS-----RDGPSRAYLGGRGVPAKSLSGFTNELLTLHG--QND
MMAR_2501|M.marinum_M SVIALRSVS-----SSGPSRAYLGGRSVPAKSLSNFTNELLALHG--QND
MUL_1685|M.ulcerans_Agy99 SVIALRSVS-----SSGPSRAYLGGRSVPAKSLSNFTNELLALHG--QND
MAV_3075|M.avium_104 SVIALRSVN-----RDGPSRAYLGGRSVPAKSLGDFTAELLTLHG--QND
MLBr_01360|M.leprae_Br4923 SVIVLRSVS-----RDGPSRSYLGGRSVPAKSLGSFTTELLALHG--QND
MAB_2361|M.abscessus_ATCC_1997 SVIAARSVT-----SDGRSRAYLGGRSVPAKSLASFTAGLLTVHG--QND
MSMEG_3808|M.smegmatis_MC2_155 TITEVYDYLRLLYARAGTPHCPVCGERIARQTPQQIVDQVLAMDEGLRFQ
:: * .:. : *. :. :: :. :*::. : :
Mflv_3507|M.gilvum_PYR-GCK QLRLMRPDEQRNALDRYA---------------------------DVTRR
Mvan_3288|M.vanbaalenii_PYR-1 QLRLMRPDEQRAALDRYA---------------------------DVATQ
TH_1548|M.thermoresistible__bu QLRLMRPDEQRAALDRFA---------------------------DLGGR
Mb1722|M.bovis_AF2122/97 QLRLMRPDEQRGALDRFA---------------------------AAGEA
Rv1696|M.tuberculosis_H37Rv QLRLMRPDEQRGALDRFA---------------------------AAGEA
MMAR_2501|M.marinum_M QLRLMRPEEQRGALDRFA---------------------------TSASA
MUL_1685|M.ulcerans_Agy99 QLRLMRPEEQRGALDRFA---------------------------TSASA
MAV_3075|M.avium_104 QLRLMRPEEQCGALDRFA---------------------------KAGPA
MLBr_01360|M.leprae_Br4923 QLRLVRPEEQRAALDRYA---------------------------AAGPS
MAB_2361|M.abscessus_ATCC_1997 QLRLMRPEQQLAALDKFASD-------------------------SIEAL
MSMEG_3808|M.smegmatis_MC2_155 VLAPVVRTRKGEFVDLFEKLNSQGYSRVRVDGVVYPLTDPPKLKKQEKHD
* : .: :* :
Mflv_3507|M.gilvum_PYR-GCK LDRYRSARDAWLEARRDLEDRRRRTRELAQEADRLHFALNEIDVVDPKPG
Mvan_3288|M.vanbaalenii_PYR-1 LHRYRTARDSWLEAKRDLDDRRRRAREMAQEADRLQFALNEIDVIDPHSG
TH_1548|M.thermoresistible__bu LQRYQLLHQQWLDARRDLADRRERARELAQEADRLTFALNEIDAVAPQPG
Mb1722|M.bovis_AF2122/97 VQRYRKLRDAWLTARRDLVDRRNRARELAQEADRLKFALNEIDTVDPQPG
Rv1696|M.tuberculosis_H37Rv VQRYRKLRDAWLTARRDLVDRRNRARELAQEADRLKFALNEIDTVDPQPG
MMAR_2501|M.marinum_M LQRYRKLRDTWLSARRDLLDRRNRARELAQEADRLQFALDEIDTVDPQPG
MUL_1685|M.ulcerans_Agy99 LQRYRKLRDTWLSARRDLLDRRNRARELAQEADRLQFALDEIDTVDPQPG
MAV_3075|M.avium_104 LERYTKLRDAWLSARRDLADRRNRMRELALEADRLTFALGEIDAVDPQPG
MLBr_01360|M.leprae_Br4923 CERYRELRDAWLLARSDLIDRRNRIRELVQEADRLKFALSEIDSVDPQPG
MAB_2361|M.abscessus_ATCC_1997 LTTYRKLREEWLAARRDLIDRTNRVRELAQEADRLGFALNEIDTVDPKPG
MSMEG_3808|M.smegmatis_MC2_155 IEVVVDRLTVKASAKQRLTDSIETALNLADGIVVLEFVDREDDHPHREQR
*: * * . ::. * *. * * .
Mflv_3507|M.gilvum_PYR-GCK EDEALVADIRRLSELDALRDAVQTARAALAGD-GDDSAHDGFSAAGAVGQ
Mvan_3288|M.vanbaalenii_PYR-1 EDDALVAEIRRLSELDALREAVQTARAALSGE-SDD----GFSAAHAVGQ
TH_1548|M.thermoresistible__bu EDDALAENIRRLSELDALREAARTARARLSGALDDDAAVDATSALDQVGQ
Mb1722|M.bovis_AF2122/97 EDVALVADIARLSELDTLREAATTARATLCG--TPDADAFDRGAVDSLGR
Rv1696|M.tuberculosis_H37Rv EDVALVADIARLSELDTLREAATTARATLCG--TPDADAFDRGAVDSLGR
MMAR_2501|M.marinum_M EDDALVADIARLSELDTLREAASAARAALSG---ASEEAFGTGAAECLGR
MUL_1685|M.ulcerans_Agy99 EDDALVADIARLSELDTLREAASAARAALSG---ASEEAFGTGAAECLGR
MAV_3075|M.avium_104 EDDALVAEIMRLSELDTLREAAAAARAALS---ADDADGSGLSAVDTLGK
MLBr_01360|M.leprae_Br4923 EDNALVADIVRLSELDMLREVAVNARAALSGA-LDDMDVSGSNAVACMGQ
MAB_2361|M.abscessus_ATCC_1997 EDDQLTADIHRLSELDALRDAAASAREALAGADADGDAHDSHSAIDRIAR
MSMEG_3808|M.smegmatis_MC2_155 FSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECTGLGIRKEVDPDLVV
. *. : :* *. . . :
Mflv_3507|M.gilvum_PYR-GCK ARSALENTDDVALKSLAEQLGAALTVVVDVSAELGHFLAELPSDASTLET
Mvan_3288|M.vanbaalenii_PYR-1 ARSALEGTDDAPLKALAEQLGSALTVLVDVSAELGGYLDELPTDTSSLET
TH_1548|M.thermoresistible__bu AKAALESTDDTALRALGARLDEALLLIGDVADELGGYLEGLPTDASTLET
Mb1722|M.bovis_AF2122/97 ARAALQSSDDAALRGLAEQVGEALTVVVDAVAELGAYLDELPADASALDA
Rv1696|M.tuberculosis_H37Rv ARAALQSSDDAALRGLAEQVGEALTVVVDAVAELGAYLDELPADASALDA
MMAR_2501|M.marinum_M ARAALESSDDATLQALAKQVSDALAVVSDAAGELGSYVDALPADASALDT
MUL_1685|M.ulcerans_Agy99 ARAALESSDDATLQALAKQVSDALAVVSDAAGELGSYVDALPADASALDT
MAV_3075|M.avium_104 ARAALESTDDPKLLALAGQIGEVLTVVVDAAGELAGFLEELPVDASALEA
MLBr_01360|M.leprae_Br4923 AKAALESTDDATLRAFADQVGEVLTVVVEVGRELGEYLEELPVDASALES
MAB_2361|M.abscessus_ATCC_1997 AKALVEATDDATLRALAPQLAEALAVVSDVSRELTAFTDELPSDASTLEE
MSMEG_3808|M.smegmatis_MC2_155 PDPELTLAEGAVAPWSVGQSAEYFTRMLAGLGEEMGFDVNTPWKKLPAKA
. . : ::. : : : * : * . . .
Mflv_3507|M.gilvum_PYR-GCK K-----------LARQAELRTLTRKYAADIDGVLAWAVEARERLSALDVS
Mvan_3288|M.vanbaalenii_PYR-1 K-----------LARQAELRTLTRKYAADVDGVLAWAGEARDRLAALDVS
TH_1548|M.thermoresistible__bu S-----------LARQSELRTLTRKYAADIDGVLQWAQQARDRLAQLDVS
Mb1722|M.bovis_AF2122/97 K-----------LARQAQLRTLTRKYAADIDGVLRWADEARARLAQLDVS
Rv1696|M.tuberculosis_H37Rv K-----------LARQAQLRTLTRKYAADIDGVLRWADEARARLAQLDVS
MMAR_2501|M.marinum_M K-----------LARQAQLRTLTRKYAADIDGVLAWAQDSRARLAALDVS
MUL_1685|M.ulcerans_Agy99 K-----------LARQAQLRTLTRKYAADIDGVLAWAQDSRARLAALDVS
MAV_3075|M.avium_104 K-----------LARQSELRGLTRKYAADIDGVLAWARESRERLAQLDVS
MLBr_01360|M.leprae_Br4923 K-----------LVRQAELCTLTRKYAADIDGVLQWARESRERLEQLDVS
MAB_2361|M.abscessus_ATCC_1997 K-----------LARQGQLRTLTRKYAADLNGVIAWAKEARTRLAEVDTS
MSMEG_3808|M.smegmatis_MC2_155 RRAILEGCDHQVHVRYKNRYGRTRSYYADFEGVMAFLQRRMEQTDSEQMK
.* : **.* **.:**: : : : .
Mflv_3507|M.gilvum_PYR-GCK E------ETLTGLQRRVDELQSAVVAAAQDVTKVRSKAAKGLSKAVTAEL
Mvan_3288|M.vanbaalenii_PYR-1 E------ETLAGLERRVEELQATVVAAAQDLTKVRVKAAKGLSKAVTAEL
TH_1548|M.thermoresistible__bu E------EALGELERRVDELQAQVVSAAMDLTKARTRAAGRLAKAVTAEL
Mb1722|M.bovis_AF2122/97 E------EGLAALERRTGELAHELGQAAVDLSTIRRKAAKRLAKEVSAEL
Rv1696|M.tuberculosis_H37Rv E------EGLAALERRTGELAHELGQAAVDLSTIRRKAAKRLAKEVSAEL
MMAR_2501|M.marinum_M E------EGLAALQRRAGELEHEVAQAAAELSKTRARAAKRLAKEVTAEL
MUL_1685|M.ulcerans_Agy99 E------EGLAALQRRAGELEHEVAQAAAELSKTRAKAAKRLAKEVTAEL
MAV_3075|M.avium_104 E------EGLSALAARVEELGRELAGAAADLSTIRRKAAKRLAKEVTAEL
MLBr_01360|M.leprae_Br4923 Y------ERLAGVESRVDELERQLSQAAVDLSKLRRDAANRLAKEVTAEL
MAB_2361|M.abscessus_ATCC_1997 E------ETLNAIAARVDRLGAELATAATGLSKARAKAAKALSKAVTAEL
MSMEG_3808|M.smegmatis_MC2_155 ERYEGFMRDIPCPECNGTRLKPEILAVTLSAGDFGAKSIAQVAELSIADC
. : . .* : .: : ::: *:
Mflv_3507|M.gilvum_PYR-GCK AGLA----------------------------------------------
Mvan_3288|M.vanbaalenii_PYR-1 SGLA----------------------------------------------
TH_1548|M.thermoresistible__bu AGLA----------------------------------------------
Mb1722|M.bovis_AF2122/97 SALA----------------------------------------------
Rv1696|M.tuberculosis_H37Rv SALA----------------------------------------------
MMAR_2501|M.marinum_M SALA----------------------------------------------
MUL_1685|M.ulcerans_Agy99 SALA----------------------------------------------
MAV_3075|M.avium_104 SGLA----------------------------------------------
MLBr_01360|M.leprae_Br4923 SALA----------------------------------------------
MAB_2361|M.abscessus_ATCC_1997 AGLA----------------------------------------------
MSMEG_3808|M.smegmatis_MC2_155 ADFLNSLTLGPREQAIAGQVLKEIQSRLGFLLDVGLDYLSLSRAAATLSG
: :
Mflv_3507|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3288|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1548|M.thermoresistible__bu --------------------------------------------------
Mb1722|M.bovis_AF2122/97 --------------------------------------------------
Rv1696|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2501|M.marinum_M --------------------------------------------------
MUL_1685|M.ulcerans_Agy99 --------------------------------------------------
MAV_3075|M.avium_104 --------------------------------------------------
MLBr_01360|M.leprae_Br4923 --------------------------------------------------
MAB_2361|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3808|M.smegmatis_MC2_155 GEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIDTLVRLRDLGN
Mflv_3507|M.gilvum_PYR-GCK -----------MSGAQFAITVAPLAARA----------DDSAPLELPSGV
Mvan_3288|M.vanbaalenii_PYR-1 -----------MAGADFAISVTPLPARA----------DDSAPLALPSGV
TH_1548|M.thermoresistible__bu -----------MADAEFTVTVGPLTARP----------DDTAPLTMPDGQ
Mb1722|M.bovis_AF2122/97 -----------MADAEFTIGVTTELADH----------GDPVALALASGE
Rv1696|M.tuberculosis_H37Rv -----------MADAEFTIGVTTELADH----------GDPVALALASGE
MMAR_2501|M.marinum_M -----------MADAEFTIAVGVEVADE----------DDAAALTLPTGE
MUL_1685|M.ulcerans_Agy99 -----------MADAEFTIVVGVEVADE----------DDAAALTLPTGE
MAV_3075|M.avium_104 -----------MADAQFSIDVSTDVVPAG------SGSDDAAVLTLPSGE
MLBr_01360|M.leprae_Br4923 -----------MVDAEFTISVTTDLIPL-------TDYKRSAAVTLPSGG
MAB_2361|M.abscessus_ATCC_1997 -----------MDRAAFSIGVEPIAARP----------DDSAPLTLPSGQ
MSMEG_3808|M.smegmatis_MC2_155 TLIVVEHDLDTIAHADWVVDIGPAAGEHGGQIVHSGTYDDLLRNPESLTG
: * : : : .
Mflv_3507|M.gilvum_PYR-GCK TVHAGRDGVDAVEFGFAAHGGSDLLPLNKSASGGELSRVMLALEVVLSAS
Mvan_3288|M.vanbaalenii_PYR-1 TVHAGHDGVDAVEFGFAAHGGSDLLPLNKSASGGELSRVMLALEVVLSAS
TH_1548|M.thermoresistible__bu TVHAGRHGVDGVEFGFTAHRGTDVLPLAKSASGGELSRVMLALEVVLAAS
Mb1722|M.bovis_AF2122/97 LARAGADGVDAVEFGFVAHRGMTVLPLAKSASGGELSRVMLSLEVVLATS
Rv1696|M.tuberculosis_H37Rv LARAGADGVDAVEFGFVAHRGMTVLPLAKSASGGELSRVMLSLEVVLATS
MMAR_2501|M.marinum_M RVRAGGDGVDLVEFGFAAHRGMTVLPLAKSASGGELSRVMLALEVVLAAS
MUL_1685|M.ulcerans_Agy99 RVRAGGDGVDLVEFGFAAHRGMTVLPLAKSASGGELSRVMLALEVVLAAS
MAV_3075|M.avium_104 RVRAGADGIDQVEFGFAAHRGMDRLPLAKSASGGELSRVMLALEVVLAAS
MLBr_01360|M.leprae_Br4923 VARAGADGVDQVEFGFAAHRGMTVLPLAKSASGGELSRVMLALEVVLATS
MAB_2361|M.abscessus_ATCC_1997 TVHAGSSGTDVVEFGFAAHRDNPMLPLAKSASGGELSRVMLALEVVLAAS
MSMEG_3808|M.smegmatis_MC2_155 AYLSGKESIEVPAIRRPVDKKRQITVVGARENNLKEIDVAFPLGVLTSVT
:* . : : .. : .. : * :.* *: :.:
Mflv_3507|M.gilvum_PYR-GCK ---TEGTTMVFDEVDAGVGG------------------------------
Mvan_3288|M.vanbaalenii_PYR-1 ---TEGTTMVFDEVDAGVGG------------------------------
TH_1548|M.thermoresistible__bu ---AEGTTMVFDEVDAGVGG------------------------------
Mb1722|M.bovis_AF2122/97 RKQAAGTTMVFDEIDAGVGG------------------------------
Rv1696|M.tuberculosis_H37Rv RKQAAGTTMVFDEIDAGVGG------------------------------
MMAR_2501|M.marinum_M ---AAGTTMVFDEVDAGVGG------------------------------
MUL_1685|M.ulcerans_Agy99 ---AAGTTMVFDEVDAGVGG------------------------------
MAV_3075|M.avium_104 RKETVGTTMVFDEVDAGVGG------------------------------
MLBr_01360|M.leprae_Br4923 ---AAGTTMVFDEVDVGVGG------------------------------
MAB_2361|M.abscessus_ATCC_1997 ---TAGTTMVFDEVDAGVGG------------------------------
MSMEG_3808|M.smegmatis_MC2_155 GVSGSGKSTMVNDILATVLANKLNGARLVPGRHTRVNGLDQLDKLVRVDQ
*.: :.::: . * .
Mflv_3507|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3288|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1548|M.thermoresistible__bu --------------------------------------------------
Mb1722|M.bovis_AF2122/97 --------------------------------------------------
Rv1696|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2501|M.marinum_M --------------------------------------------------
MUL_1685|M.ulcerans_Agy99 --------------------------------------------------
MAV_3075|M.avium_104 --------------------------------------------------
MLBr_01360|M.leprae_Br4923 --------------------------------------------------
MAB_2361|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3808|M.smegmatis_MC2_155 SPIGRTPRSNPATYTGVFDKIRSLFAATTEAKVRGYQPGRFSFNVKGGRC
Mflv_3507|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3288|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1548|M.thermoresistible__bu --------------------------------------------------
Mb1722|M.bovis_AF2122/97 --------------------------------------------------
Rv1696|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2501|M.marinum_M --------------------------------------------------
MUL_1685|M.ulcerans_Agy99 --------------------------------------------------
MAV_3075|M.avium_104 --------------------------------------------------
MLBr_01360|M.leprae_Br4923 --------------------------------------------------
MAB_2361|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3808|M.smegmatis_MC2_155 EACSGDGTIKIEMNFLPDVYVPCEVCHGARYNRETLEVHYKGKTISEVLD
Mflv_3507|M.gilvum_PYR-GCK --------------------RAAVQIGRRLARLARTHQVIVVTHLPQVAA
Mvan_3288|M.vanbaalenii_PYR-1 --------------------RAAVQIGRRLARLARTHQVIVVTHLPQVAA
TH_1548|M.thermoresistible__bu --------------------RAAVQIGRRLARLARTHQVIVVTHLPQVAA
Mb1722|M.bovis_AF2122/97 --------------------WAAVQIGRRLARLARTHQVIVVTHLPQVAA
Rv1696|M.tuberculosis_H37Rv --------------------WAAVQIGRRLARLARTHQVIVVTHLPQVAA
MMAR_2501|M.marinum_M --------------------WAAVQIGRRLARLARTHQVIVVTHLPQVAA
MUL_1685|M.ulcerans_Agy99 --------------------WAAVQIGRRLARLARTHQVIVVTHLPQVAA
MAV_3075|M.avium_104 --------------------RAAVQIGRRLARLARTHQVIVVTHLPQVAA
MLBr_01360|M.leprae_Br4923 --------------------RAAVQIGRRLARLARTHQVIVVTHLPQVAA
MAB_2361|M.abscessus_ATCC_1997 --------------------RAAVQIGRRLAKLAHTHQVIVVTHLPQVAA
MSMEG_3808|M.smegmatis_MC2_155 MSIEEATEFFEPISSIHRYLKTLVDVGLGYVRLGQPAPTLSGGEAQRVKL
: *::* .:*.:. .: . :*
Mflv_3507|M.gilvum_PYR-GCK YADVHLVVEGVGNDGAGSGDG--------------KKARAKSSGVRRLDD
Mvan_3288|M.vanbaalenii_PYR-1 YADVHLVVEGV----AGRDDG--------------RKARAKSSGVRRLDD
TH_1548|M.thermoresistible__bu YADIHLVVDSG-----GRDAD--------------NRG---ASRVQRLDE
Mb1722|M.bovis_AF2122/97 YADVHLMVQRTG--------------------------RDGASGVRRLTS
Rv1696|M.tuberculosis_H37Rv YADVHLMVQRTG--------------------------RDGASGVRRLTS
MMAR_2501|M.marinum_M YADVHLMVHGT---------------------------KGGASEVRRLSD
MUL_1685|M.ulcerans_Agy99 YADVHLMVHGT---------------------------KGGASEVRRLSD
MAV_3075|M.avium_104 YADVHLVVHGAG--------------------------PRGTSVVRRVTG
MLBr_01360|M.leprae_Br4923 YADVHLVVHSAG--------------------------LDGASDVRRLAG
MAB_2361|M.abscessus_ATCC_1997 FADIHLTVDRVN---------------------------SKGSGVRRLDD
MSMEG_3808|M.smegmatis_MC2_155 AAELQKRSTGRTIYILDEPTTGLHFEDIRKLLKVINGLVDKGNTVIVIEH
*::: . * :
Mflv_3507|M.gilvum_PYR-GCK DDRVAELARMLAGLGE---SDSGRAHAR-------------------DLL
Mvan_3288|M.vanbaalenii_PYR-1 DDRVAELARMLAGLGE---SDSGRAHAR-------------------ELL
TH_1548|M.thermoresistible__bu EGRVAELARMLAGLGE---TDSGRAHAR-------------------ELL
Mb1722|M.bovis_AF2122/97 EDRVAELARMLAGLGD---SDSGRAHAR-------------------ELL
Rv1696|M.tuberculosis_H37Rv EDRVAELARMLAGLGD---SDSGRAHAR-------------------ELL
MMAR_2501|M.marinum_M DERVGELARMLAGLGE---SDSGRAHAR-------------------ELL
MUL_1685|M.ulcerans_Agy99 DERVGELARMLAGLGE---SDSGRAHAR-------------------ELL
MAV_3075|M.avium_104 DERVAELARMLAGLGE---SDSGRAHAR-------------------ELL
MLBr_01360|M.leprae_Br4923 DDRVAELARMLAGLGE---SDSGRAHAR-------------------ELL
MAB_2361|M.abscessus_ATCC_1997 EDRVAELARMLAGLGD---SDTGRAHAR-------------------ELL
MSMEG_3808|M.smegmatis_MC2_155 NLDVIKTSDWIIDMGPEGGAGGGTVVAQGTPEDVAANPDSYTGKFLAELL
: * : : : .:* :. * . *: :**
Mflv_3507|M.gilvum_PYR-GCK DAARKDRAAE----
Mvan_3288|M.vanbaalenii_PYR-1 DAARAERAAAD---
TH_1548|M.thermoresistible__bu EAAQRERAEVSV--
Mb1722|M.bovis_AF2122/97 ETAQNDELT-----
Rv1696|M.tuberculosis_H37Rv ETAQNDELT-----
MMAR_2501|M.marinum_M DTARNDKD------
MUL_1685|M.ulcerans_Agy99 DTARNDKD------
MAV_3075|M.avium_104 DAAQKDEI------
MLBr_01360|M.leprae_Br4923 DAARKDKS------
MAB_2361|M.abscessus_ATCC_1997 DAARAERT------
MSMEG_3808|M.smegmatis_MC2_155 DVPTPKRKRRKVSA
:.. ..