For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTWDRSVDMLIAGSGGGGMVAGLAALDSGLEPLIVEKQSLVGGSTGLSGGIVWLPNNPLMRAEGVADSHQ DGLAYLADVVGDIGAASSPERREMFLTAGHEMLNFLTRRGVALIRCAGWSDYYPNHKGGNASGRAVEAVP FDAAKLGSWSGRVQPPLARNYGYVVLTNELRSVQYFNRSARTFAVAARVFARTAAARMRRRQLLTNGASL IAHMLKALLDLSDGQPPLWTDAALTELVVEDGRVVGACIVRDGRACAVEARKGVLLAAGGFGHNKQMRRR YSGDQPNEGQWSIANAGDTGEVLEAAMRLGAKTDLMDEAWWLPSVFIANGGAVAASLGSGRQRPGAIYVD STGRRFCNESNSYVEVGKAMYANKAVPCWMIFDDGYVRRYVTSANPLKTLRRHQPLPAELIESGAVKRAA TIAGLAGETDLPADELARSVERFNRFAAKGLDPDFGRGQSAYNDCLGDPGYRPNAAIGPLDTAPYYATRV FPADVGTCGGVITNEYAQVLDQHDRVIEGLYATGNTTATVMGRTYPGAGASIANSMVFGYVAARHAAGRR IAGTPPRAIAGVPPRRVCD
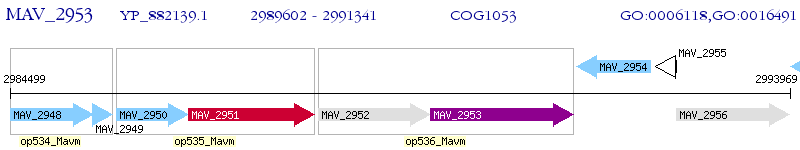
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2953 | - | - | 100% (579) | 3-ketosteroid-delta-1-dehydrogenase |
| M. avium 104 | MAV_0624 | - | e-105 | 38.38% (568) | 3-ketosteroid-delta-1-dehydrogenase |
| M. avium 104 | MAV_0880 | - | 2e-99 | 38.78% (575) | 3-ketosteroid-delta-1-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3567 | - | 1e-105 | 38.62% (567) | 3-ketosteroid-delta-1-dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1519 | - | 1e-104 | 38.39% (573) | 3-ketosteroid-delta-1-dehydrogenase |
| M. tuberculosis H37Rv | Rv3537 | - | 1e-105 | 38.62% (567) | 3-ketosteroid-delta-1-dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0622c | - | 1e-104 | 39.24% (576) | 3-ketosteroid-delta-1-dehydrogenase |
| M. marinum M | MMAR_5024 | - | 1e-105 | 38.42% (570) | dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5941 | - | 1e-105 | 38.81% (572) | 3-ketosteroid-delta-1-dehydrogenase |
| M. thermoresistible (build 8) | TH_4337 | - | 1e-101 | 37.82% (587) | PUTATIVE fumarate reductase/succinate dehydrogenase |
| M. ulcerans Agy99 | MUL_4098 | - | 1e-104 | 38.88% (571) | 3-ketosteroid-delta-1-dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5237 | - | 1e-104 | 38.45% (567) | 3-ketosteroid-delta-1-dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1519|M.gilvum_PYR-GCK ----MAQQEYDVVVVGSGAAGMVAALTAAHQGLSTIVVEKAPHYGGSTAR
Mvan_5237|M.vanbaalenii_PYR-1 ----MTEQEYDVVVVGSGAAGMVAALTAAHQGLSTIVVEKAPHYGGSTAR
MSMEG_5941|M.smegmatis_MC2_155 ----MTGQEYDVVVVGSGAAGMVAALTAAHQGLSTVVVEKAPHYGGSTAR
Mb3567|M.bovis_AF2122/97 ----MTVQEFDVVVVGSGAAGMVAALVAAHRGLSTVVVEKAPHYGGSTAR
Rv3537|M.tuberculosis_H37Rv ----MTVQEFDVVVVGSGAAGMVAALVAAHRGLSTVVVEKAPHYGGSTAR
MMAR_5024|M.marinum_M -MFYMTSQDYDVVVVGSGGAGMVAALAAAHKGLSTVVIEKAAHYGGSTAR
MUL_4098|M.ulcerans_Agy99 ----MTSQDYDVVVVGSGGAGMVAALAAAHKGLSTVVIEKAAHYGGSTAR
MAB_0622c|M.abscessus_ATCC_199 ----MTEQEYDVVVVGSGGAGMVAALTAAHNGLSTVLIEKAPHFGGSTAR
TH_4337|M.thermoresistible__bu MTVDTHDQTVDVLVVGSGGGGMTAALTADAAGLDTLLVEKSPQFGGSTAL
MAV_2953|M.avium_104 ---MTWDRSVDMLIAGSGGGGMVAGLAALDSGLEPLIVEKQSLVGGSTGL
: *:::.***..**.*.*.* **..:::** . ****.
Mflv_1519|M.gilvum_PYR-GCK SGGGVWIPNNEVLKRDGVKDTTEAARTYLHSIVG----DAVPAEKIDTYL
Mvan_5237|M.vanbaalenii_PYR-1 SGGGVWIPNNEVLKRDGVKDTADAARTYLHAIIG----DVVPAEKIDTYL
MSMEG_5941|M.smegmatis_MC2_155 SGGGVWIPNNEILKRDGVKDTPDEARKYLHAIIG----DVVPAEKIDTYL
Mb3567|M.bovis_AF2122/97 SGGGVWIPNNEVLKRRGVRDTPEAARTYLHGIVG----EIVEPERIDAYL
Rv3537|M.tuberculosis_H37Rv SGGGVWIPNNEVLKRRGVRDTPEAARTYLHGIVG----EIVEPERIDAYL
MMAR_5024|M.marinum_M SGGGVWIPNNQILKRARVSDTTEAARTYLHGIIGDSSVSGVEPERIDTYL
MUL_4098|M.ulcerans_Agy99 SGGGVWIPNNQILKRARVSDTTEAARTYLHGIIGDSSVSGVEPERIDTYL
MAB_0622c|M.abscessus_ATCC_199 SGGGVWIPNNEVLKKARQTDTPEAAREYLYSIIG----DVVPREKIDTYL
TH_4337|M.thermoresistible__bu SGGGIWVPGAPAQRREGYSPDPDDVFEYLRQITG----GLVSDARLRTYV
MAV_2953|M.avium_104 SGGIVWLPNNPLMRAEGVADSHQDGLAYLADVVGDIG-AASSPERREMFL
*** :*:*. : : ** : * : ::
Mflv_1519|M.gilvum_PYR-GCK DRGPEMLSFVLKHSP-LKLCWVPGYSDYYPETPGGKPTGRSVEPKPFNAK
Mvan_5237|M.vanbaalenii_PYR-1 QRGPEMLSFVLKHSP-LKLCWVPGYSDYYPETPGGKATGRSVEPKPFDAK
MSMEG_5941|M.smegmatis_MC2_155 DRGPEMLSFVLKHSP-LKLCWVPGYSDYYPETPGGKPTGRSVEPKPFDAN
Mb3567|M.bovis_AF2122/97 DRGPEMLSFVLKHTP-LKMCWVPGYSDYYPEAPGGRPGGRSIEPKPFNAR
Rv3537|M.tuberculosis_H37Rv DRGPEMLSFVLKHTP-LKMCWVPGYSDYYPEAPGGRPGGRSIEPKPFNAR
MMAR_5024|M.marinum_M ERGPEMLSFVLEHTP-LKMCWVPGYSDYYPEAPGGRSGGRSIEPKPFDAR
MUL_4098|M.ulcerans_Agy99 ERGPEMLSFVLEHTP-LKMCWVPGYSDYYSEAPGGRSGGRSIEPKPFDAR
MAB_0622c|M.abscessus_ATCC_199 DRGPEMLSFVLANSP-LKLDWVPNYSDYYPEAPGGRAAGRSVEPKPFDAK
TH_4337|M.thermoresistible__bu DEAPKMMEFLEQLSPWCEFVWKPGYADYYPELPGGSPLGSTINVPAIDLR
MAV_2953|M.avium_104 TAGHEMLNFLTRRGV--ALIRCAGWSDYYPNHKGGNASGRAVEAVPFDAA
. :*:.*: : ..::***.: ** . * ::: .::
Mflv_1519|M.gilvum_PYR-GCK KLGPDEKGLEPPYGKVPLNMVVMQQDYVRLNQLKRHPRGVLRSLKVGVRS
Mvan_5237|M.vanbaalenii_PYR-1 KLGEDLPGLEPPYGKVPMNMVVMQQDYVRLNQLKRHPRGVLRSLRVGIRS
MSMEG_5941|M.smegmatis_MC2_155 KLGPDLKGLEPPYGKVPMNMVVMQQDYVRLNQLKRHPRGVLRSLKVGIRA
Mb3567|M.bovis_AF2122/97 KLGADMAGLEPAYGKVPLNVVVMQQDYVRLNQLKRHPRGVLRSMKVGART
Rv3537|M.tuberculosis_H37Rv KLGADMAGLEPAYGKVPLNVVVMQQDYVRLNQLKRHPRGVLRSMKVGART
MMAR_5024|M.marinum_M KLGADMAELEPAYGKIPLNVVVMQQDYVRLNQLKRHPRGVLRSMKVGART
MUL_4098|M.ulcerans_Agy99 KLGADMAELEPAYGKIPLNVVVMQQDYVRLNQLKRHPRGVLRSMKVGART
MAB_0622c|M.abscessus_ATCC_199 KLGDELPNLEPPYGKVPLNVVVRQQDYVRLNQLKRHPRGVLRSLRVGVRT
TH_4337|M.thermoresistible__bu RLGDEEARLLQPLALAPRGIWFAPKDLRLFYQVRQNWRGKAVLVKLIWRM
MAV_2953|M.avium_104 KLGSWSGRVQPPLARN-YGYVVLTNELRSVQYFNRSARTFAVAARVFART
:** : . . . . :: . ..: * :: *
Mflv_1519|M.gilvum_PYR-GCK VWANATGKNLVGMGRALIAPLR---IGLQKAGVPVLLNTALTDLHVE-DG
Mvan_5237|M.vanbaalenii_PYR-1 VWANTTGKNLVGMGRALIAPLR---IGLREAGVPVRLNTAMTDLYVE-DG
MSMEG_5941|M.smegmatis_MC2_155 TWGKVSGKNLVGMGRALIAPLR---IGLREAGVPVLLNTALTDLYVE-DG
Mb3567|M.bovis_AF2122/97 MWAKATGKNLVGMGRALIGPLR---IGLQRAGVPVELNTAFTDLFVE-NG
Rv3537|M.tuberculosis_H37Rv MWAKATGKNLVGMGRALIGPLR---IGLQRAGVPVELNTAFTDLFVE-NG
MMAR_5024|M.marinum_M MWAKATGKNLVGMGRALIGPLR---IGLQRKGVPVQLNTALTDLYLE-DG
MUL_4098|M.ulcerans_Agy99 MWAKATGKNLVGMGRALIGPLR---IGLQRKGVPVQLNTALTDLYLE-DG
MAB_0622c|M.abscessus_ATCC_199 FWAQGTGKKLVGMGRALSAALR---IGLRDAGVPVKLNTALTDLYIE-DG
TH_4337|M.thermoresistible__bu FRARVFGDRMAAIGQSLAARLR---LAMRQQGVPLWLNAPMTELITDADG
MAV_2953|M.avium_104 AAARMRRRQLLTNGASLIAHMLKALLDLSDGQPPLWTDAALTELVVE-DG
.. .: * :* . : : : *: ::.:*:* : :*
Mflv_1519|M.gilvum_PYR-GCK VVTGIYVVATGDSVNAEPQLIRARRAVILGSGGFEHNEQMRVKY-QRAPI
Mvan_5237|M.vanbaalenii_PYR-1 VVRGIYVRDTTAPESAEPQLIRARRGVILGSGGFEHNEQMRVKY-QRAPI
MSMEG_5941|M.smegmatis_MC2_155 RVLGIYVRDTTAGDDAEPRLIRARHGVILGSGGFEHNEQMRVKY-QRAPI
Mb3567|M.bovis_AF2122/97 VVSGVYVRDSHEAESAEPQLIRARRGVILACGGFEHNEQMRIKY-QRAPI
Rv3537|M.tuberculosis_H37Rv VVSGVYVRDSHEAESAEPQLIRARRGVILACGGFEHNEQMRIKY-QRAPI
MMAR_5024|M.marinum_M MVRGIYVRSTDAQESEEPQLIRARKGVILACGGFDHNEQMRVKY-QRAPI
MUL_4098|M.ulcerans_Agy99 VVRGIYVRSTDAHESEEPQLIRARKGVILACGGFDHNEQMRVKY-QRAPI
MAB_0622c|M.abscessus_ATCC_199 TVRGVYVRETGAPEASEPTLIRGRRGVILASGGFEHNEQMRVKY-QRAPI
TH_4337|M.thermoresistible__bu AVVGAVVEKDGRAVR-----IRARHGVILAAGGFDHDMDRRRE--HLPLL
MAV_2953|M.avium_104 RVVGACIVRDGRACA-----VEARKGVLLAAGGFGHNKQMRRRYSGDQPN
* * : :..*:.*:*..*** *: : * .
Mflv_1519|M.gilvum_PYR-GCK TTEWTVGAVANTGDGILAAEKLGAALEIMEDAWWGPTVPLEG--APWFAL
Mvan_5237|M.vanbaalenii_PYR-1 TTEWTVGAKANTGDGILAAEKLGAALDVMEDAWWGPTVPLVD--APWFAL
MSMEG_5941|M.smegmatis_MC2_155 TTEWTVGAVANTGDGIVAAEKLGAALELMEDAWWGPTVPLVG--APWFAL
Mb3567|M.bovis_AF2122/97 TTEWTVGASANTGDGILAAEKLGAALDLMDDAWWGPTVPLVG--KPWFAL
Rv3537|M.tuberculosis_H37Rv TTEWTVGASANTGDGILAAEKLGAALDLMDDAWWGPTVPLVG--KPWFAL
MMAR_5024|M.marinum_M TTDWTVGAKANTGDGIVAAEKLGAALDLMEDAWWGPTVPLVG--RPWFAL
MUL_4098|M.ulcerans_Agy99 TTDWTVGAKANTGDGIVAAEKLGAALDLMEDAWWGPTVPLVG--RPWFAL
MAB_0622c|M.abscessus_ATCC_199 TTEWTVGAKANTGDGILAAEKLGAALEFMEDSWWGPTVPLVG--RPWFAL
TH_4337|M.thermoresistible__bu EKDWSFGNPAATGDGIRAGEKVGGATELLDEAWWFPAICWPDG-RLQFML
MAV_2953|M.avium_104 EGQWSIANAGDTGEVLEAAMRLGAKTDLMDEAWWLPSVFIANGGAVAASL
:*:.. . **: : *. ::*. :.::::** *:: . *
Mflv_1519|M.gilvum_PYR-GCK SE-RNSPGSIIVNMAGKRFMNESMPYVEACHHMYGGQYGQGPGPGEN---
Mvan_5237|M.vanbaalenii_PYR-1 SE-RNSPGSIIVNMAGKRFMNESMPYVEACHHMYGGQYGQGPGPGEN---
MSMEG_5941|M.smegmatis_MC2_155 SE-RNSPGSIIVNMSGKRFMNESMPYVEACHHMYGGQYGQGPGPGEN---
Mb3567|M.bovis_AF2122/97 SE-RNSPGSIIVNMSGKRFMNESMPYVEACHHMYGGEHGQGPGPGEN---
Rv3537|M.tuberculosis_H37Rv SE-RNSPGSIIVNMSGKRFMNESMPYVEACHHMYGGEHGQGPGPGEN---
MMAR_5024|M.marinum_M SE-RNSPGSIIVNMSGQRFMNESMPYVEACHHMYGGQYGQGPGPGEN---
MUL_4098|M.ulcerans_Agy99 SE-RNSPGSIIVNMSGQRFMNESMPYVEACHHMYGGQYGQGPGPGEN---
MAB_0622c|M.abscessus_ATCC_199 SE-RNSPGSIIVNSSGKRFMNESLPYVEATHRMYGGQYGQGPGPGEN---
TH_4337|M.thermoresistible__bu NE-RMMPSQFIVNGAGRRFINEAAPYMDFAHAMIEGERAYRDSHGENGTA
MAV_2953|M.avium_104 GSGRQRPGAIYVDSTGRRFCNESNSYVEVGKAMYANKA------------
.. * *. : *: :*:** **: .*:: : * .:
Mflv_1519|M.gilvum_PYR-GCK --IPAWLIFDQQYRDRYIFAG-----------LQPGQRIPSKWLESGVIV
Mvan_5237|M.vanbaalenii_PYR-1 --IPAWLVFDQRYRDRYIFAG-----------LQPGQRIPKKWLESGVIV
MSMEG_5941|M.smegmatis_MC2_155 --IPAWLIFDQQYRDRYIFAG-----------LQPGQRIPSKWLESGVIV
Mb3567|M.bovis_AF2122/97 --IPAWLVFDQRYRDRYIFAG-----------LQPGQRIPSRWLDSGVIV
Rv3537|M.tuberculosis_H37Rv --IPAWLVFDQRYRDRYIFAG-----------LQPGQRIPSRWLDSGVIV
MMAR_5024|M.marinum_M --IPAWLVFDQRYRDRYIFAG-----------LQPGQRIPRKWLDSGVIV
MUL_4098|M.ulcerans_Agy99 --IPAWLVFDQRYRDRYIFAG-----------LQPGQRIPRKWLDSGVIV
MAB_0622c|M.abscessus_ATCC_199 --LPAWLVFDQEYRNRYIFAG-----------LQPGQKIPSKWIESGVIV
TH_4337|M.thermoresistible__bu QHIPCWLITDIRSFRRYVVAGHLPIPKIPFAPVPTGRKVPREWLESGVVH
MAV_2953|M.avium_104 --VPCWMIFDDGYVRRYVTSAN------PLKTLRRHQPLPAELIESGAVK
:*.*:: * **: :. : : :* . ::**.:
Mflv_1519|M.gilvum_PYR-GCK KADTLDELAARTGLPTDAFKATVDRFNGFARSGRDEDFKRGESAYDRYYG
Mvan_5237|M.vanbaalenii_PYR-1 KADTLEELAAKTGLPVDAFKTTVQRFNGFARSGVDEDFKRGESAYDRYYG
MSMEG_5941|M.smegmatis_MC2_155 KADTLVELAEKTGLPADQFTSTIERFNGFARSGVDEDFHRGESAYDRYYG
Mb3567|M.bovis_AF2122/97 QADTLAELAGKAGLPADELTATVQRFNAFARSGVDEDYHRGESAYDRYYG
Rv3537|M.tuberculosis_H37Rv QADTLAELAGKAGLPADELTATVQRFNAFARSGVDEDYHRGESAYDRYYG
MMAR_5024|M.marinum_M MADTLEELAAKAGLPVDELTSTVSRFNGFARSGVDEDYHRGESAYDRYYG
MUL_4098|M.ulcerans_Agy99 MADTLEELAAKAGLPVDELTSTVSRFNGFARSGVDEDYHRGESAYDRYYG
MAB_0622c|M.abscessus_ATCC_199 KADTIEELAKKANLPVEELRATIERFNGFARSGVDEDFGRGKSAYDRYYG
TH_4337|M.thermoresistible__bu EGHSWEELAAKINVPADQLRATAERFNMLAAKGHDDDFNRGDSAYDNYYG
MAV_2953|M.avium_104 RAATIAGLAGETDLPADELARSVERFNRFAAKGLDPDFGRGQSAYNDCLG
. : ** . .:*.: : : .*** :* .* * *: **.***: *
Mflv_1519|M.gilvum_PYR-GCK DPTNKPNPNLGEITQGPFYAAKMVPGDLGTKGGIRTDVHGRALRDDGSVI
Mvan_5237|M.vanbaalenii_PYR-1 DPTNKPNPNLGEISQGPFYAAKMVPGDLGTKGGIVTDVQGRALRDDGSII
MSMEG_5941|M.smegmatis_MC2_155 DPTNKPNPNLGEIKHGPFYAAKMVPGDLGTKGGIRTDVRGRALRDDDSVI
Mb3567|M.bovis_AF2122/97 DPSNKPNPNLGEVGHPPYYGAKMVPGDLGTKGGIRTDVNGRALRDDGSII
Rv3537|M.tuberculosis_H37Rv DPSNKPNPNLGEVGHPPYYGAKMVPGDLGTKGGIRTDVNGRALRDDGSII
MMAR_5024|M.marinum_M DPTNKPNPNLGEINHAPYYAAKMVPGDLGTKGGIRTDVNGRALRDDGSII
MUL_4098|M.ulcerans_Agy99 DPTNKPNPNLGEINHAPYYAAKMVPGDLGTKGGIRTDVNGRALRDDGSII
MAB_0622c|M.abscessus_ATCC_199 DPTNKPNPNLGALTKAPFYAAKMVPGDLGTKGGVRTDVKGRVLRDDNSVI
TH_4337|M.thermoresistible__bu DPT-LPNPNLHPLGRPPYYAFQIILGDLGTSGGLRTDEHARVLRVDDSVV
MAV_2953|M.avium_104 DPGYRPNAAIGPLDTAPYYATRVFPADVGTCGGVITNEYAQVLDQHDRVI
** **. : : *:*. ::. .*:** **: *: .:.* .. ::
Mflv_1519|M.gilvum_PYR-GCK EGLYAAGNVSSPVMGHTYPGPGGTIGPAMTFGYLAALHIAGAAPAATSSS
Mvan_5237|M.vanbaalenii_PYR-1 EGLYAAGNASAPVMGHTYPGPGGTIGPAMTFGYLAALDIARKD-------
MSMEG_5941|M.smegmatis_MC2_155 EGLYAAGNVSSPVMGHTYPGPGGTIGPAMTFGYLAALDIAATAAASRKG-
Mb3567|M.bovis_AF2122/97 DGLYAAGNVSAPVMGHTYPGPGGTIGPAMTFGYLAALHIADQAGKR----
Rv3537|M.tuberculosis_H37Rv DGLYAAGNVSAPVMGHTYPGPGGTIGPAMTFGYLAALHIADQAGKR----
MMAR_5024|M.marinum_M DGLYAAGNVSAPVMGHTYPGPGGTIGPAMTFGYLAALHIAGDR-------
MUL_4098|M.ulcerans_Agy99 DGLYAAGNVSAPVMGHTYPGPGGTIGPAMTFGYLAALHIAGDR-------
MAB_0622c|M.abscessus_ATCC_199 EGLYAAGNVSTPVMGHTYAGPGATIGPAMTWGYLAALDIAGK--------
TH_4337|M.thermoresistible__bu PGLYAVGNNSAAVMGRSYAGAGATIGPAMTFGYVAARHIAERAAPATTSS
MAV_2953|M.avium_104 EGLYATGNTTATVMGRTYPGAGASIANSMVFGYVAARHAAGRRIAGTPPR
****.** ::.***::*.*.*.:*. :*.:**:** . *
Mflv_1519|M.gilvum_PYR-GCK GKG---------
Mvan_5237|M.vanbaalenii_PYR-1 ------------
MSMEG_5941|M.smegmatis_MC2_155 ------------
Mb3567|M.bovis_AF2122/97 ------------
Rv3537|M.tuberculosis_H37Rv ------------
MMAR_5024|M.marinum_M ------------
MUL_4098|M.ulcerans_Agy99 ------------
MAB_0622c|M.abscessus_ATCC_199 ------------
TH_4337|M.thermoresistible__bu PTRPSTSKSNVS
MAV_2953|M.avium_104 AIAGVPPRRVCD