For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGVGATPYYKRGGSLPKSITELAGEAILAACDDAGLPVTDVDGFAYYSGASAGYTEKMDTADFMETLGIP EVRFTAALTSGGGGSAGAIGLARAAIVAGDASVVVTVMALQQSKQRLGSVFSAMDPDPINSFLQPSGLFG PGQLMSVLARRHMHLYGTRREAFAEIALSTRANAFNRPKAMHRQPLTLDDYFNARMIAEPLCLYDFCQET DGAVAVITTSMDRARDLRQPPVPVVAAAHGGVRDWGRAFAWMGMPDEYFASSGNAPIAKRLYEQAAISAA DIDVALLYDHFSPMVLMQLEDYGFCGKGEGGAFVESGAIRYDGGSIPVNTHGGQLSEAYIIGMTHIMEGV EQMRGTAINQVRDAELALVTGGPASLPVSGLILGRAA
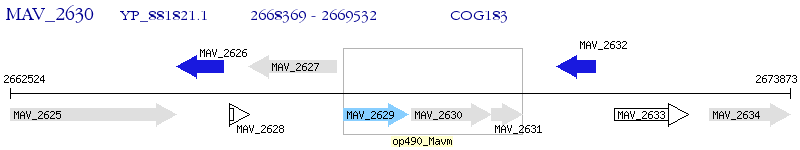
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2630 | - | - | 100% (387) | thiolase |
| M. avium 104 | MAV_1918 | - | 0.0 | 93.80% (387) | thiolase |
| M. avium 104 | MAV_0477 | - | 8e-85 | 44.90% (392) | lipid-transfer protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3570c | ltp2 | 1e-74 | 42.08% (404) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_1513 | - | 2e-75 | 41.88% (394) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv3540c | ltp2 | 1e-74 | 42.08% (404) | lipid-transfer protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0618 | - | 2e-75 | 41.12% (394) | lipid-transfer protein |
| M. marinum M | MMAR_5027 | ltp2_1 | 2e-74 | 41.34% (404) | acetyl-CoA acetyltransferase (PaaJ-like), Ltp2_1 |
| M. smegmatis MC2 155 | MSMEG_5990 | - | 3e-75 | 41.37% (394) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_4616 | - | 0.0 | 90.70% (387) | PUTATIVE putative nonspecific lipid-transfer protein |
| M. ulcerans Agy99 | MUL_4101 | ltp2_1 | 1e-74 | 41.34% (404) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_5253 | - | 8e-76 | 41.88% (394) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1513|M.gilvum_PYR-GCK ---MLSGKAAIAGIGATDFSKNSG---RSELRLAAEAVLDALDDAGLTTA
Mvan_5253|M.vanbaalenii_PYR-1 ---MLSGKAAIAGIGATDFSKNSG---RSELRLAAEAVLDALDDAGLTPS
MAB_0618|M.abscessus_ATCC_1997 -MTGLSGRSAIAGIGATDFSKKSG---RSELRLAAEAVLDALDDAGLSPS
MSMEG_5990|M.smegmatis_MC2_155 -MTGLSGKAAIAGIGATDFSKNSG---RSELRLAAECVLDALDDAGLEPS
Mb3570c|M.bovis_AF2122/97 ---MLSGQAAIVGIGATDFSKNSG---RSELRLAAEAVLDALADAGLSPT
Rv3540c|M.tuberculosis_H37Rv ---MLSGQAAIVGIGATDFSKNSG---RSELRLAAEAVLDALADAGLSPT
MMAR_5027|M.marinum_M ---MLSGKAAIVGIGATDFSKNSG---RSELRLAAEAVRDALDDAGLSPA
MUL_4101|M.ulcerans_Agy99 ---MLSGKAAIVGIGATDFSKNSG---RSELRLAAEAVRDALDDAGLSPA
MAV_2630|M.avium_104 -----------MGVGATPYYKRGGSLPKSITELAGEAILAACDDAGLPVT
TH_4616|M.thermoresistible__bu VDSNGLRDVAIVGVGATPFYKRGGSLPKTIVELAGEAIIAACEDAGLPVH
*:*** : *..* :: .**.*.: * ****
Mflv_1513|M.gilvum_PYR-GCK DVDGLVTFS------MDSNLETAVARSTGIGELKFFSQIGYGGGAAAATV
Mvan_5253|M.vanbaalenii_PYR-1 DVDGLVTFT------MDSNLETAVARSTGIGDLTFFSQIGYGGGAAAATV
MAB_0618|M.abscessus_ATCC_1997 DVDGLVTFT------MDSNLETAVARATGIGELKFFSQIGYGGGAAAATV
MSMEG_5990|M.smegmatis_MC2_155 DVDGLVTFT------MDSNQETAVARSTGIGELKFFSQIGYGGGAAAATV
Mb3570c|M.bovis_AF2122/97 DVDGLTTFT------MDTNTEIAVARAAGIGELTFFSKIHYGGGAACATV
Rv3540c|M.tuberculosis_H37Rv DVDGLTTFT------MDTNTEIAVARAAGIGELTFFSKIHYGGGAACATV
MMAR_5027|M.marinum_M DVDGLTTFT------MDTNTEIAVARAAGIGELKFFSKIHYGGGAACATV
MUL_4101|M.ulcerans_Agy99 DVDGLTTFT------MDTNTEIAVARAAGIGELKFFSKIHYGGGAACATV
MAV_2630|M.avium_104 DVDGFAYYSGASAGYTEKMDTADFMETLGIPEVRFTAALTSGGGGSAGAI
TH_4616|M.thermoresistible__bu DIDGFAYYSGANAGYTDKMDTADFMETLGIPEVRFTAALTSGGGGSAGAI
*:**:. :: :. . .: ** :: * : : ***.:..::
Mflv_1513|M.gilvum_PYR-GCK QQAALAVATGVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYSWS
Mvan_5253|M.vanbaalenii_PYR-1 QQAALAVATGVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYSWS
MAB_0618|M.abscessus_ATCC_1997 QQAALAVAAGVAEVVVAYRAFNERSEFRFGQVMTGLSSTADSRGVEYSWS
MSMEG_5990|M.smegmatis_MC2_155 QQAALAVATGVAEVVVAYRAFNERSEFRFGQVMTGLTTNADSRGVEYSWS
Mb3570c|M.bovis_AF2122/97 QHAAMAVATGVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSFS
Rv3540c|M.tuberculosis_H37Rv QHAAMAVATGVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSFS
MMAR_5027|M.marinum_M QHAAMAVATGIADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSFS
MUL_4101|M.ulcerans_Agy99 QHAAMAVATGIADVVVAYRAFNERSDMRFGQVQTRLTENADSTGVDNSFS
MAV_2630|M.avium_104 GLARAAIVAGDASVVVTVMALQQ-SKQRLGSVFSAMDPDP-----INSFL
TH_4616|M.thermoresistible__bu GLARAAIVAGDASVVVTVMALQQ-AKQRLGSVFSAMERDP-----INSFL
* *:.:* *.***: *::: : *:*.* : : . *:
Mflv_1513|M.gilvum_PYR-GCK YPHGLSTPAASVAMIAQRYMHEFGATSADFGVISVADRKHAANNPKAHFY
Mvan_5253|M.vanbaalenii_PYR-1 YPHGLSTPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAHFY
MAB_0618|M.abscessus_ATCC_1997 YPHGLSTPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAFFY
MSMEG_5990|M.smegmatis_MC2_155 YPHGLSTPAASVAMIARRYMHEYGATSADFGVVSVADRKHAANNPKAHFY
Mb3570c|M.bovis_AF2122/97 YPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYFY
Rv3540c|M.tuberculosis_H37Rv YPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYFY
MMAR_5027|M.marinum_M YPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAYFY
MUL_4101|M.ulcerans_Agy99 YPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAYFY
MAV_2630|M.avium_104 QPSGLFGPGQLMSVLARRHMHLYGTRREAFAEIALSTRANAFNRPKAMHR
TH_4616|M.thermoresistible__bu QPSGLFGPGQLMSVLARRHMHLYGTRREAFAEIAISTRANALNRPNAMYR
* ** *. ::::*:*:** *: *. :::: * :* ..*:* .
Mflv_1513|M.gilvum_PYR-GCK GKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERARDLKHRPA
Mvan_5253|M.vanbaalenii_PYR-1 GKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLRHRPA
MAB_0618|M.abscessus_ATCC_1997 GKPITIEDHQNSRMIADPVRLLDCCQESDGGVAIVVTTPERAKDLKNRPV
MSMEG_5990|M.smegmatis_MC2_155 GKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLKHRPV
Mb3570c|M.bovis_AF2122/97 GKPITIEDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQRPV
Rv3540c|M.tuberculosis_H37Rv GKPITIEDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQRPV
MMAR_5027|M.marinum_M QKPITIDDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQRPA
MUL_4101|M.ulcerans_Agy99 QKPITIDDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQRPA
MAV_2630|M.avium_104 -QPLTLDDYFNARMIAEPLCLYDFCQETDGAVAVITTSMDRARDLRQPPV
TH_4616|M.thermoresistible__bu -EPLTLENYFNARMIAEPLCLYDFCLETDGAVAVITTSMDRARDLRQPPV
:*:*:::: *:* **:*: * * * *:**.**::.*: **:**:: *.
Mflv_1513|M.gilvum_PYR-GCK IIEAAAQGAGADQFTMYSYYREELG-LPEMG--LVGRQLWAQSGLTPQDI
Mvan_5253|M.vanbaalenii_PYR-1 VIEAAAQGAGADQFTMYSYYRDELG-LPEMG--LVGRQLWEQSGLSPEDI
MAB_0618|M.abscessus_ATCC_1997 IIEAAAQGSGEDQFTMYSYYRDELG-LPEMG--LVGRQLWDQSGLTPNDI
MSMEG_5990|M.smegmatis_MC2_155 TIEAAAQGSGADQFTMYSYYRDELG-LPEMG--LVGRQLWQQSGLTPDDI
Mb3570c|M.bovis_AF2122/97 VIEAAAQGCSPDQYTMVSYYRPELDGLPEMG--LVGRQLWAQSGLTPADV
Rv3540c|M.tuberculosis_H37Rv VIEAAAQGCSPDQYTMVSYYRPELDGLPEMG--LVGRQLWAQSGLTPADV
MMAR_5027|M.marinum_M IIEAASQGSSPDQYTMVSYYRPELDGLPEMG--LVGQQLWQQSGLKPTDI
MUL_4101|M.ulcerans_Agy99 IIEAASQGSSPDQYTMVSYYRPELDGLPEMG--LVGQQLWQQSGLKPTDI
MAV_2630|M.avium_104 PVVAAAHGGVRDWGRAFAWMGMPDEYFASSGNAPIAKRLYEQAAISAADI
TH_4616|M.thermoresistible__bu PVIAAAHGGVRDWGRAFAWMGMADDYFASSGNRPIADRLYRQADITADDI
: **::* * :: :.. * :. :*: *: :.. *:
Mflv_1513|M.gilvum_PYR-GCK QTAVLYDHFTPYTLIQLEELGFCGKGEAKDFIAGGAIEI-GGRLPINTHG
Mvan_5253|M.vanbaalenii_PYR-1 QTAILYDHFTPYTLIQLEELGFCGKGEAKDFIAGGAIEI-GGRLPINTHG
MAB_0618|M.abscessus_ATCC_1997 QTAILYDHFTPYTLLQLEELGFCGKGEAKDFIANGAIEL-GGKLPINTHG
MSMEG_5990|M.smegmatis_MC2_155 QTAILYDHFTPYTLLQLEELGFCGKGEAKDFIAGGAIEI-GGKLPINTHG
Mb3570c|M.bovis_AF2122/97 QTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEV-GGRLPINTHG
Rv3540c|M.tuberculosis_H37Rv QTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEV-GGRLPINTHG
MMAR_5027|M.marinum_M QTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEL-GGRLPINTHG
MUL_4101|M.ulcerans_Agy99 QTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEL-GGRLPINTHG
MAV_2630|M.avium_104 DVALLYDHFSPMVLMQLEDYGFCGKGEGGAFVESGAIRYDGGSIPVNTHG
TH_4616|M.thermoresistible__bu DVALLYDHFTPMVLMQLEDYGFCGKGEGGAFVESGAIRYDGGAIPVNPHG
:.*:*****:* .*:***: *******. *: .***. ** :*:*.**
Mflv_1513|M.gilvum_PYR-GCK GQLGEAYIHGMNGIAEGVRQLRGTSVNQVPDVEHVLVTAG-TGVPTSGLI
Mvan_5253|M.vanbaalenii_PYR-1 GQLGEAYIHGMNGIAEGVRQLRGTSVNQVPDVEHVLVTAG-TGVPTSGLI
MAB_0618|M.abscessus_ATCC_1997 GQLGEAYIHGMNGIAEGVRQLRGTSVNQVPNVEHVLVTAG-TGVPTSGLI
MSMEG_5990|M.smegmatis_MC2_155 GQLGEAYIHGMNGIAEGVRQLRGTSVNQVEGVEHVLVTAG-TGVPTSGLI
Mb3570c|M.bovis_AF2122/97 GQLGEAYIHGMNGIAEGVRQLRGTSVNPVAGVEHVLVTAG-TGVPTSGLI
Rv3540c|M.tuberculosis_H37Rv GQLGEAYIHGMNGIAEGVRQLRGTSVNPVAGVEHVLVTAG-TGVPTSGLI
MMAR_5027|M.marinum_M GQLGEAYIHGMNGIAEGVRQLRGTSVNPVPDVEHVLVTAG-TGVPTSGLI
MUL_4101|M.ulcerans_Agy99 GQLGEAYIHGMNGIAEGVRQLRGTSVNPVPDVEHVLVTAG-TGVPTSGLI
MAV_2630|M.avium_104 GQLSEAYIIGMTHIMEGVEQMRGTAINQVRDAELALVTGGPASLPVSGLI
TH_4616|M.thermoresistible__bu GQLSEAYIVGMTHIMEGVEQMRGTAINQVPNAELALVTGGPASLPVSGLI
***.**** **. * ***.*:***::* * ..* .***.* :.:*.****
Mflv_1513|M.gilvum_PYR-GCK LG---
Mvan_5253|M.vanbaalenii_PYR-1 LG---
MAB_0618|M.abscessus_ATCC_1997 LG---
MSMEG_5990|M.smegmatis_MC2_155 LG---
Mb3570c|M.bovis_AF2122/97 LG---
Rv3540c|M.tuberculosis_H37Rv LG---
MMAR_5027|M.marinum_M LG---
MUL_4101|M.ulcerans_Agy99 LG---
MAV_2630|M.avium_104 LGRAA
TH_4616|M.thermoresistible__bu LGKAA
**