For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSNTSGLRDVAIVGVGATDYYKRGGSLPKSITELAGEAILAACEDAGIPVTDIDGFAYYSGASAGYTEKM DTADFVEALGIPEVRFTAALTSGGGGSAGAIGLARAAIVAGDASVVVTVMALQQAKQRLGSVFSAMDPDP INSFLQPSGLFGPGQLMSVLARRHMHLYGTRREAFAEIALSTRTNALNRPKAMHRQPLTLDDYFNARMIA EPLCLYDFCQETDGAVAVITTSMERAKDLKQPPVPVVAAAHGGVRDWGRAFAWMGMPDEYFASSGNAPIA KRLYEQAGVGPADIDVALLYDHFSPMVLMQLEDYGFCDKGEGGAFVESGAIRFGSGSIPVNTHGGQLSEA YIIGMTHIVEGVEQMRGTAINQVKDAELALITGGPASLPVSGLILGKAA
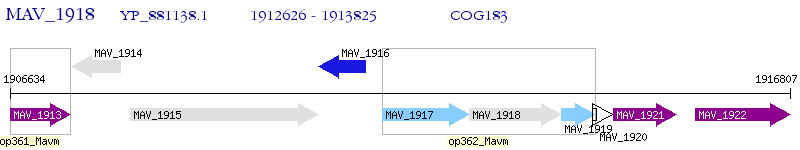
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1918 | - | - | 100% (399) | thiolase |
| M. avium 104 | MAV_2630 | - | 0.0 | 93.80% (387) | thiolase |
| M. avium 104 | MAV_0477 | - | 2e-85 | 44.92% (394) | lipid-transfer protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3570c | ltp2 | 5e-77 | 42.12% (406) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_1513 | - | 4e-77 | 41.31% (397) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv3540c | ltp2 | 5e-77 | 42.12% (406) | lipid-transfer protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0618 | - | 2e-78 | 41.31% (397) | lipid-transfer protein |
| M. marinum M | MMAR_5027 | ltp2_1 | 2e-76 | 41.13% (406) | acetyl-CoA acetyltransferase (PaaJ-like), Ltp2_1 |
| M. smegmatis MC2 155 | MSMEG_5990 | - | 1e-77 | 41.31% (397) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_4616 | - | 0.0 | 87.41% (397) | PUTATIVE putative nonspecific lipid-transfer protein |
| M. ulcerans Agy99 | MUL_4101 | ltp2_1 | 9e-77 | 41.13% (406) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_5253 | - | 3e-77 | 41.06% (397) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0618|M.abscessus_ATCC_1997 MTGLSGR--SAIAGIGATDFSKKSG---RSELRLAAEAVLDALDDAGLSP
MSMEG_5990|M.smegmatis_MC2_155 MTGLSGK--AAIAGIGATDFSKNSG---RSELRLAAECVLDALDDAGLEP
Mflv_1513|M.gilvum_PYR-GCK --MLSGK--AAIAGIGATDFSKNSG---RSELRLAAEAVLDALDDAGLTT
Mvan_5253|M.vanbaalenii_PYR-1 --MLSGK--AAIAGIGATDFSKNSG---RSELRLAAEAVLDALDDAGLTP
Mb3570c|M.bovis_AF2122/97 --MLSGQ--AAIVGIGATDFSKNSG---RSELRLAAEAVLDALADAGLSP
Rv3540c|M.tuberculosis_H37Rv --MLSGQ--AAIVGIGATDFSKNSG---RSELRLAAEAVLDALADAGLSP
MMAR_5027|M.marinum_M --MLSGK--AAIVGIGATDFSKNSG---RSELRLAAEAVRDALDDAGLSP
MUL_4101|M.ulcerans_Agy99 --MLSGK--AAIVGIGATDFSKNSG---RSELRLAAEAVRDALDDAGLSP
MAV_1918|M.avium_104 MSNTSGLRDVAIVGVGATDYYKRGGSLPKSITELAGEAILAACEDAGIPV
TH_4616|M.thermoresistible__bu -VDSNGLRDVAIVGVGATPFYKRGGSLPKTIVELAGEAIIAACEDAGLPV
.* **.*:*** : *..* :: .**.*.: * ***:
MAB_0618|M.abscessus_ATCC_1997 SDVDGLVTFT------MDSNLETAVARATGIGELKFFSQIGYGGGAAAAT
MSMEG_5990|M.smegmatis_MC2_155 SDVDGLVTFT------MDSNQETAVARSTGIGELKFFSQIGYGGGAAAAT
Mflv_1513|M.gilvum_PYR-GCK ADVDGLVTFS------MDSNLETAVARSTGIGELKFFSQIGYGGGAAAAT
Mvan_5253|M.vanbaalenii_PYR-1 SDVDGLVTFT------MDSNLETAVARSTGIGDLTFFSQIGYGGGAAAAT
Mb3570c|M.bovis_AF2122/97 TDVDGLTTFT------MDTNTEIAVARAAGIGELTFFSKIHYGGGAACAT
Rv3540c|M.tuberculosis_H37Rv TDVDGLTTFT------MDTNTEIAVARAAGIGELTFFSKIHYGGGAACAT
MMAR_5027|M.marinum_M ADVDGLTTFT------MDTNTEIAVARAAGIGELKFFSKIHYGGGAACAT
MUL_4101|M.ulcerans_Agy99 ADVDGLTTFT------MDTNTEIAVARAAGIGELKFFSKIHYGGGAACAT
MAV_1918|M.avium_104 TDIDGFAYYSGASAGYTEKMDTADFVEALGIPEVRFTAALTSGGGGSAGA
TH_4616|M.thermoresistible__bu HDIDGFAYYSGANAGYTDKMDTADFMETLGIPEVRFTAALTSGGGGSAGA
*:**:. :: :. . .: ** :: * : : ***.:..:
MAB_0618|M.abscessus_ATCC_1997 VQQAALAVAAGVAEVVVAYRAFNERSEFRFGQVMTGLSSTADSRGVEYSW
MSMEG_5990|M.smegmatis_MC2_155 VQQAALAVATGVAEVVVAYRAFNERSEFRFGQVMTGLTTNADSRGVEYSW
Mflv_1513|M.gilvum_PYR-GCK VQQAALAVATGVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYSW
Mvan_5253|M.vanbaalenii_PYR-1 VQQAALAVATGVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYSW
Mb3570c|M.bovis_AF2122/97 VQHAAMAVATGVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSF
Rv3540c|M.tuberculosis_H37Rv VQHAAMAVATGVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSF
MMAR_5027|M.marinum_M VQHAAMAVATGIADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSF
MUL_4101|M.ulcerans_Agy99 VQHAAMAVATGIADVVVAYRAFNERSDMRFGQVQTRLTENADSTGVDNSF
MAV_1918|M.avium_104 IGLARAAIVAGDASVVVTVMALQQ-AKQRLGSVFSAMDPDP-----INSF
TH_4616|M.thermoresistible__bu IGLARAAIVAGDASVVVTVMALQQ-AKQRLGSVFSAMERDP-----INSF
: * *:.:* *.***: *::: : *:*.* : : . *:
MAB_0618|M.abscessus_ATCC_1997 SYPHGLSTPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAFF
MSMEG_5990|M.smegmatis_MC2_155 SYPHGLSTPAASVAMIARRYMHEYGATSADFGVVSVADRKHAANNPKAHF
Mflv_1513|M.gilvum_PYR-GCK SYPHGLSTPAASVAMIAQRYMHEFGATSADFGVISVADRKHAANNPKAHF
Mvan_5253|M.vanbaalenii_PYR-1 SYPHGLSTPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAHF
Mb3570c|M.bovis_AF2122/97 SYPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYF
Rv3540c|M.tuberculosis_H37Rv SYPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYF
MMAR_5027|M.marinum_M SYPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAYF
MUL_4101|M.ulcerans_Agy99 SYPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAYF
MAV_1918|M.avium_104 LQPSGLFGPGQLMSVLARRHMHLYGTRREAFAEIALSTRTNALNRPKAMH
TH_4616|M.thermoresistible__bu LQPSGLFGPGQLMSVLARRHMHLYGTRREAFAEIAISTRANALNRPNAMY
* ** *. ::::*:*:** *: *. :::: * :* ..*:* .
MAB_0618|M.abscessus_ATCC_1997 YGKPITIEDHQNSRMIADPVRLLDCCQESDGGVAIVVTTPERAKDLKNRP
MSMEG_5990|M.smegmatis_MC2_155 YGKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLKHRP
Mflv_1513|M.gilvum_PYR-GCK YGKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERARDLKHRP
Mvan_5253|M.vanbaalenii_PYR-1 YGKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLRHRP
Mb3570c|M.bovis_AF2122/97 YGKPITIEDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQRP
Rv3540c|M.tuberculosis_H37Rv YGKPITIEDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQRP
MMAR_5027|M.marinum_M YQKPITIDDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQRP
MUL_4101|M.ulcerans_Agy99 YQKPITIDDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQRP
MAV_1918|M.avium_104 R-QPLTLDDYFNARMIAEPLCLYDFCQETDGAVAVITTSMERAKDLKQPP
TH_4616|M.thermoresistible__bu R-EPLTLENYFNARMIAEPLCLYDFCLETDGAVAVITTSMDRARDLRQPP
:*:*:::: *:* **:*: * * * *:**.**::.*: **:**:: *
MAB_0618|M.abscessus_ATCC_1997 VIIEAAAQGSGEDQFTMYSYYRDELG-LPEMG--LVGRQLWDQSGLTPND
MSMEG_5990|M.smegmatis_MC2_155 VTIEAAAQGSGADQFTMYSYYRDELG-LPEMG--LVGRQLWQQSGLTPDD
Mflv_1513|M.gilvum_PYR-GCK AIIEAAAQGAGADQFTMYSYYREELG-LPEMG--LVGRQLWAQSGLTPQD
Mvan_5253|M.vanbaalenii_PYR-1 AVIEAAAQGAGADQFTMYSYYRDELG-LPEMG--LVGRQLWEQSGLSPED
Mb3570c|M.bovis_AF2122/97 VVIEAAAQGCSPDQYTMVSYYRPELDGLPEMG--LVGRQLWAQSGLTPAD
Rv3540c|M.tuberculosis_H37Rv VVIEAAAQGCSPDQYTMVSYYRPELDGLPEMG--LVGRQLWAQSGLTPAD
MMAR_5027|M.marinum_M AIIEAASQGSSPDQYTMVSYYRPELDGLPEMG--LVGQQLWQQSGLKPTD
MUL_4101|M.ulcerans_Agy99 AIIEAASQGSSPDQYTMVSYYRPELDGLPEMG--LVGQQLWQQSGLKPTD
MAV_1918|M.avium_104 VPVVAAAHGGVRDWGRAFAWMGMPDEYFASSGNAPIAKRLYEQAGVGPAD
TH_4616|M.thermoresistible__bu VPVIAAAHGGVRDWGRAFAWMGMADDYFASSGNRPIADRLYRQADITADD
. : **::* * :: :.. * :. :*: *:.: . *
MAB_0618|M.abscessus_ATCC_1997 IQTAILYDHFTPYTLLQLEELGFCGKGEAKDFIANGAIELG-GKLPINTH
MSMEG_5990|M.smegmatis_MC2_155 IQTAILYDHFTPYTLLQLEELGFCGKGEAKDFIAGGAIEIG-GKLPINTH
Mflv_1513|M.gilvum_PYR-GCK IQTAVLYDHFTPYTLIQLEELGFCGKGEAKDFIAGGAIEIG-GRLPINTH
Mvan_5253|M.vanbaalenii_PYR-1 IQTAILYDHFTPYTLIQLEELGFCGKGEAKDFIAGGAIEIG-GRLPINTH
Mb3570c|M.bovis_AF2122/97 VQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEVG-GRLPINTH
Rv3540c|M.tuberculosis_H37Rv VQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEVG-GRLPINTH
MMAR_5027|M.marinum_M IQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIELG-GRLPINTH
MUL_4101|M.ulcerans_Agy99 IQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIELG-GRLPINTH
MAV_1918|M.avium_104 IDVALLYDHFSPMVLMQLEDYGFCDKGEGGAFVESGAIRFGSGSIPVNTH
TH_4616|M.thermoresistible__bu IDVALLYDHFTPMVLMQLEDYGFCGKGEGGAFVESGAIRYDGGAIPVNPH
::.*:*****:* .*:***: ***.***. *: .***. . * :*:*.*
MAB_0618|M.abscessus_ATCC_1997 GGQLGEAYIHGMNGIAEGVRQLRGTSVNQVPNVEHVLVTAG-TGVPTSGL
MSMEG_5990|M.smegmatis_MC2_155 GGQLGEAYIHGMNGIAEGVRQLRGTSVNQVEGVEHVLVTAG-TGVPTSGL
Mflv_1513|M.gilvum_PYR-GCK GGQLGEAYIHGMNGIAEGVRQLRGTSVNQVPDVEHVLVTAG-TGVPTSGL
Mvan_5253|M.vanbaalenii_PYR-1 GGQLGEAYIHGMNGIAEGVRQLRGTSVNQVPDVEHVLVTAG-TGVPTSGL
Mb3570c|M.bovis_AF2122/97 GGQLGEAYIHGMNGIAEGVRQLRGTSVNPVAGVEHVLVTAG-TGVPTSGL
Rv3540c|M.tuberculosis_H37Rv GGQLGEAYIHGMNGIAEGVRQLRGTSVNPVAGVEHVLVTAG-TGVPTSGL
MMAR_5027|M.marinum_M GGQLGEAYIHGMNGIAEGVRQLRGTSVNPVPDVEHVLVTAG-TGVPTSGL
MUL_4101|M.ulcerans_Agy99 GGQLGEAYIHGMNGIAEGVRQLRGTSVNPVPDVEHVLVTAG-TGVPTSGL
MAV_1918|M.avium_104 GGQLSEAYIIGMTHIVEGVEQMRGTAINQVKDAELALITGGPASLPVSGL
TH_4616|M.thermoresistible__bu GGQLSEAYIVGMTHIMEGVEQMRGTAINQVPNAELALVTGGPASLPVSGL
****.**** **. * ***.*:***::* * ..* .*:*.* :.:*.***
MAB_0618|M.abscessus_ATCC_1997 ILG---
MSMEG_5990|M.smegmatis_MC2_155 ILG---
Mflv_1513|M.gilvum_PYR-GCK ILG---
Mvan_5253|M.vanbaalenii_PYR-1 ILG---
Mb3570c|M.bovis_AF2122/97 ILG---
Rv3540c|M.tuberculosis_H37Rv ILG---
MMAR_5027|M.marinum_M ILG---
MUL_4101|M.ulcerans_Agy99 ILG---
MAV_1918|M.avium_104 ILGKAA
TH_4616|M.thermoresistible__bu ILGKAA
***