For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVESPGTPRLAVAGKPLREVGSYFALTLDIFVQLVHPPFAWREFVHQAWFVARVSIAPTILLSIPFNALS VFIINVLLVEIGAADASGAGAALASVAYIGPITTVLVVAGTGATAICADLGARTIREELDAMRVMGINPV QRLLVPRVLALCLNGLLLNSINTIVGLVGSFFFSVYFQHVTPGAWAASLTLLVKIADVAIAFFKAALFGL VAGTIACYKGVTVGGGPQGVGNAVNETVVYTFMALFVINVVLVAVGTKVTM
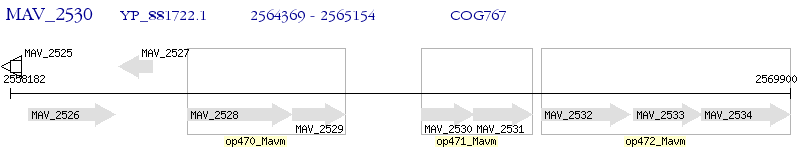
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2530 | - | - | 100% (261) | TrnB1 protein |
| M. avium 104 | MAV_2059 | - | 7e-84 | 58.54% (246) | TrnB1 protein |
| M. avium 104 | MAV_0946 | - | 3e-82 | 56.70% (261) | TrnB1 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3531c | yrbE4A | 1e-79 | 57.55% (245) | integral membrane protein YrbE4a |
| M. gilvum PYR-GCK | Mflv_2099 | - | 4e-93 | 66.00% (250) | hypothetical protein Mflv_2099 |
| M. tuberculosis H37Rv | Rv1964 | yrbE3A | 6e-82 | 60.74% (242) | integral membrane protein YrbE3A |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 2e-72 | 53.50% (243) | hypothetical protein MLBr_02587 |
| M. abscessus ATCC 19977 | MAB_4155c | - | 5e-77 | 56.15% (244) | hypothetical protein MAB_4155c |
| M. marinum M | MMAR_4049 | yrbE3A_1 | 8e-93 | 66.27% (252) | integral membrane protein YrbE3A_1 |
| M. smegmatis MC2 155 | MSMEG_0344 | - | 2e-91 | 63.10% (252) | hypothetical protein MSMEG_0344 |
| M. thermoresistible (build 8) | TH_0179 | - | 3e-90 | 63.97% (247) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE4A |
| M. ulcerans Agy99 | MUL_3916 | yrbE3A_1 | 5e-93 | 66.27% (252) | integral membrane protein YrbE3A_1 |
| M. vanbaalenii PYR-1 | Mvan_0920 | - | 4e-93 | 67.21% (247) | hypothetical protein Mvan_0920 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2530|M.avium_104 -MVESP---GTPRLAVAGKPLREVGSYFALTLDIFVQLVHPPFAWREFVH
Mvan_0920|M.vanbaalenii_PYR-1 ----------MPATESFARPMRAVGGFFATAVDTAVLAPQPPFAWREFIF
MMAR_4049|M.marinum_M -MVAPV---------AVAKPLRAFGGFYSMALDTFVAMVHPPFAWREYIA
MUL_3916|M.ulcerans_Agy99 -MVTPV---------AVAKPLRAFGGFYSMALDTFVAMVHPPFAWREYIA
Mflv_2099|M.gilvum_PYR-GCK -MVASG---------VIAKPMRAFGGFFSMALDTFVAMVKPPFAWREFIS
MSMEG_0344|M.smegmatis_MC2_155 -MIALG---------PAAKPVGALGGFFAMTLDVFITIARPPFAWREYLN
Rv1964|M.tuberculosis_H37Rv -MVIVADKAAGRVADPVLRPVGALGDFFAMTLDTSVCMFKPPFAWREYLL
TH_0179|M.thermoresistible__bu -MVLAP--------QPVTEPVRAVGGFFAMSLDTFVQMFRPPFAWREYVH
Mb3531c|M.bovis_AF2122/97 ------------MIQQLAVPARAVGGFFEMSMDTARAAFRRPFQFREFLD
MAB_4155c|M.abscessus_ATCC_199 ------------MKQQLAAPARAVGGFVSMSLATFRNIFRRPFQGQEFLD
MLBr_02587|M.leprae_Br4923 MTTETRSSLVEYAHTKLQTPLVLVGGFFRMLVLTGKALFRRPFQWREFIL
* .*.: : : ** :*::
MAV_2530|M.avium_104 QAWFVARVSIAPTILLSIPFNALSVFIINVLLVEIGAADASGAGAALASV
Mvan_0920|M.vanbaalenii_PYR-1 QMWFVARVSIFPTVMLSIPFTVLSVFIFNILLVEFGAADFSGAGAAFGAV
MMAR_4049|M.marinum_M QAWFVARVSLLPTLMLTIPYTVLLTFIENILLTEIGAADFSGTGAALGSV
MUL_3916|M.ulcerans_Agy99 QAWFVARVSLLPTLMLTIPYTVLLTFIENILLTEIGAADFSGTGAALGSV
Mflv_2099|M.gilvum_PYR-GCK QSWFVARVSIVPTLMLTIPYTVLLTFTFNILLTEFGAADFSGTGAALGTV
MSMEG_0344|M.smegmatis_MC2_155 QTWFVARVSLVPALMLTVPYTVLLVFTFNILLIEFGAADFSGTGAAIGTV
Rv1964|M.tuberculosis_H37Rv QCWFVARVSTLPGVLMTIPWAVISGFLFNVLLTDIGAADFSGTGCAIFTV
TH_0179|M.thermoresistible__bu QSWFVARVSVVPALLLTMPYSVLLVFTFNVLLAEFGAADYSGTGAAIGTV
Mb3531c|M.bovis_AF2122/97 QTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGAADLSGAGTAFGTI
MAB_4155c|M.abscessus_ATCC_199 QTWFIARVSLLPTLLVAIPFTVLVAFTLNILLREIGAADLSGAATAFGTV
MLBr_02587|M.leprae_Br4923 QCWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQFSAADLSGAGAAIGAV
* *:: **: * :::::* .: * *:** ::.*** **:. *: ::
MAV_2530|M.avium_104 AYIGPITTVLVVAGTGATAICADLGARTIREELDAMRVMGINPVQRLLVP
Mvan_0920|M.vanbaalenii_PYR-1 TQIGPLVTVLVVAGAAATAMCADLGARTIREELDAMRVMGINPLQALVVP
MMAR_4049|M.marinum_M RQIGPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGVDPIQALVVP
MUL_3916|M.ulcerans_Agy99 RQIGPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGVDPIQALVVP
Mflv_2099|M.gilvum_PYR-GCK RQIGPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGVNPIQALVVP
MSMEG_0344|M.smegmatis_MC2_155 SQIGPIVTVLVVAGAGSTAMCADLGARTIREELDAMRVMGIDPIQALVVP
Rv1964|M.tuberculosis_H37Rv NQSAPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGINPIQALAAP
TH_0179|M.thermoresistible__bu NQIGPIVTVLVVSGAGATAMCADLGARTIREELDALRVMGINPIQALVVP
Mb3531c|M.bovis_AF2122/97 TQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMRVLGIDPIQRLVVP
MAB_4155c|M.abscessus_ATCC_199 TQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMQVLGIDPIQRLVVP
MLBr_02587|M.leprae_Br4923 TQLGPLTTVLVVAGAGSTSICADLGARTIREEIDAMEVLGIDPIHRLVVP
.*:.*****:*:.:*::************:**:.*:*::*:: * .*
MAV_2530|M.avium_104 RVLALCLNGLLLNSINTIVGLVGSFFFSVYFQHVTPGAWAASLTLLVKIA
Mvan_0920|M.vanbaalenii_PYR-1 RVLATVVVALLLNSVVSVVGVVGGFFFSVFVQHVTPGAFVAGMTLFTGLP
MMAR_4049|M.marinum_M RVLAATTVSLALSASVILVGLSGAYFFCVFIQHVSPGAFAAGLTLIIGTT
MUL_3916|M.ulcerans_Agy99 RVLAATTVSLALSASVILVGLSGAYFFCVFIQHVSPGAFAAGLTLIIGTT
Mflv_2099|M.gilvum_PYR-GCK RVLAATLVSLALSATVILVGLAGAYFFCVYIQNVSPGAFAAGLTLIIGAT
MSMEG_0344|M.smegmatis_MC2_155 RVLAATTVALALSATVIIVGLAGGFVFAVFIQHVPPGSFVAGLTVLTHGA
Rv1964|M.tuberculosis_H37Rv RVLAATTVSLALNSVVTATGLIGAFFCSVFLMHVSAGAWVTGLTTLTHTV
TH_0179|M.thermoresistible__bu RVLAATTVSLALASTVILVGLVGAFFFVVYVQHVSAGAFVTGLTLMTGAA
Mb3531c|M.bovis_AF2122/97 RVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPGAFINGLTVLTGLR
MAB_4155c|M.abscessus_ATCC_199 RVLASTFVALLLNGLVCAIGMVGGYVFSVFLQGVNPGAFINGLTVLTGLG
MLBr_02587|M.leprae_Br4923 RVLAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGGAYLATLTTITGLP
**** . * . *: *.: *:. * *:: :* :
MAV_2530|M.avium_104 DVAIAFFKAALFGLVAGTIACYKGVTVGGGPQGVGNAVNETVVYTFMALF
Mvan_0920|M.vanbaalenii_PYR-1 EIIISLTKASLFGLVAALIGCYKGISVGGGPVGVGTAVNETVVFSFMALF
MMAR_4049|M.marinum_M DVVIALVKAALFGLSAGLIACYKGISVGGGPAGVGNAVNETVVFTFMALF
MUL_3916|M.ulcerans_Agy99 DVVIALVKAALFGLSAGLIACYKGISVGGGPAGVGNAVNETVVFTFMALF
Mflv_2099|M.gilvum_PYR-GCK DVIIALVKAALFGLSAGMIACYKGISVGGGPAGVGNAVNETVVFTFMALF
MSMEG_0344|M.smegmatis_MC2_155 DVAVALAKAALFGLSAGLIACYKGISVGGGPAGVGNAVNETVVFTFMALF
Rv1964|M.tuberculosis_H37Rv DVVISMIKATLFGLMAGLIACYKGMSVGGGPAGVGRAVNETVVFAFIVLF
TH_0179|M.thermoresistible__bu DVIVSLVKATLFGMAAGLIACYKGISVGGGPAGVGNAVNETVVFSFMVLF
Mb3531c|M.bovis_AF2122/97 ELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNAVNETVVYAFICLF
MAB_4155c|M.abscessus_ATCC_199 ELMISEVKAFLFGIFAGLVGCYRGLTVKGGPKGVGDAVNETVVYAFICLF
MLBr_02587|M.leprae_Br4923 EVVIATVKAATFGLIAGLVGCYRGLIVRGGSNGLGAAVNETVVLCVVALY
:: :: ** **: *. :.**:*: * **. *:* ******* .: *:
MAV_2530|M.avium_104 VINVVLVAVGTKVTM--
Mvan_0920|M.vanbaalenii_PYR-1 VINVVVTAVGVKATM--
MMAR_4049|M.marinum_M AINIVATAVAIKVTL--
MUL_3916|M.ulcerans_Agy99 AINIVATAVAIKVTL--
Mflv_2099|M.gilvum_PYR-GCK AINIVATAVAVKVTM--
MSMEG_0344|M.smegmatis_MC2_155 AINVIATAVGVKITVV-
Rv1964|M.tuberculosis_H37Rv VINIVVTAVGIPFMVS-
TH_0179|M.thermoresistible__bu AINIVVTAVGIQFTV--
Mb3531c|M.bovis_AF2122/97 VINVVMTAIGVRISAQ-
MAB_4155c|M.abscessus_ATCC_199 VINVVMTAIGVRVLVKH
MLBr_02587|M.leprae_Br4923 AVNVVLTTIGVRFGTGH
.:*:: .::.