For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRLGTKALAVVAGVAVVAAAVGIGWWLLRPDDDTITVTAQFDSASGLYEGNVVAVLGMPVGKITKINPRG GYVEVEFTVDRGVKVPANAQAVTVSTSILTDRQIELTPPYRGGPVLGNHDTIGLPRTKTPVEFSSVLNVL DKVTKSLEGDGHGGGPIADVLGGGAEVVNGNGEKIKAALGELSKALRLSSDGGAATREQITTIVKNLSTL FDAVASNDTKLREFASTIHQVSQIMADEDLGSGNTGHKLDQLIQRAGDLLDANRDNIKHAVLSGNDTLKT VTDQRRDLAELLDLAPLVADNAYNMIDRANGSVRARFLTDRLLFDSQYTKEICNLMGLRQLGCSTGTIQD FGPDFGLTYVLDGLAAMGQK*
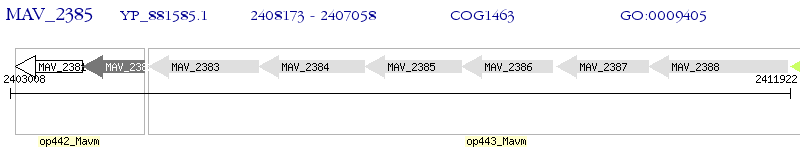
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2385 | - | - | 100% (371) | virulence factor Mce family protein |
| M. avium 104 | MAV_0105 | - | e-128 | 61.10% (365) | virulence factor Mce family protein |
| M. avium 104 | MAV_0660 | - | 2e-42 | 35.05% (311) | virulence factor mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3526c | mce4D | 2e-43 | 34.41% (311) | MCE-family protein MCE4D |
| M. gilvum PYR-GCK | Mflv_0007 | - | 1e-134 | 63.81% (362) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv3496c | mce4D | 2e-43 | 34.41% (311) | MCE-family protein MCE4D |
| M. leprae Br4923 | MLBr_02592 | mce1D | 3e-32 | 28.89% (315) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_1007c | - | 1e-144 | 68.58% (366) | putative MCE family protein |
| M. marinum M | MMAR_3867 | mce5D | 1e-170 | 78.44% (371) | MCE-family protein Mce5D |
| M. smegmatis MC2 155 | MSMEG_1146 | - | 1e-130 | 62.19% (365) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_3076 | - | 1e-129 | 61.04% (367) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_4916 | mce6D | 1e-135 | 64.19% (363) | MCE-family protein Mce6D |
| M. vanbaalenii PYR-1 | Mvan_0962 | - | 1e-134 | 63.98% (372) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2385|M.avium_104 -------MTRLGT---KALAVVAGVAVVAA-AVGIGWWLLRPDDDTITVT
MMAR_3867|M.marinum_M -------MTTSKV---KILAVVAAVLAVVA-LAGAGWWRMGADDDPMTVT
MAB_1007c|M.abscessus_ATCC_199 -------MNSKGAGKGRAIAVIATVAAVAAAIVGACVYYVKSQLDHITLT
MUL_4916|M.ulcerans_Agy99 -------MTHRHN----LIRVGVGLVVVAA-LACAGWYGLARDRGPIVVT
Mflv_0007|M.gilvum_PYR-GCK --------MSGRA---KVTALVVAGLVAVVAVAAIGWSFLSDRLDAITVT
Mvan_0962|M.vanbaalenii_PYR-1 --------MGTRS---KLVSLAVAAVAVVVVVAAVGWSYLGDRMDTISVT
MSMEG_1146|M.smegmatis_MC2_155 -------MKLLRN---KVAAALLAAVVLGAVVLAVGGAYVKQQMGAITVT
TH_3076|M.thermoresistible__bu ------MMRDVRT---RVIALIAAGLVAVAAVAVVAVKVARDQLDTITVT
Mb3526c|M.bovis_AF2122/97 -----MMGRVAMLTGSRGLRYATVIALVAALVGGVY--VLSSTGNKRTIV
Rv3496c|M.tuberculosis_H37Rv -----MMGRVAMLTGSRGLRYATVIALVAALVGGVY--VLSSTGNKRTIV
MLBr_02592|M.leprae_Br4923 MSTIFDVRKLKLLRLSRAPVIVGSLVVVLALVVGFVGWRLYQKLTNNTVV
. :.
MAV_2385|M.avium_104 AQFDSASGLYEGNVVAVLGMPVGKITKINPRGGYVEVEFTVDRGVKVPAN
MMAR_3867|M.marinum_M AQFDSASGLYEGNVVAVLGMPVGQVTKIRPKGGYVEVEFTIDRKVKIPAD
MAB_1007c|M.abscessus_ATCC_199 AQFDNASGLYESNVVAVLGMPVGKITKITPQSGYVDVEFTVDKDVKVPVD
MUL_4916|M.ulcerans_Agy99 AQFDSAAGLYEGNAVAVLGIPVGHITKIVAKSGYAEVEFTVDRAVQVPAD
Mflv_0007|M.gilvum_PYR-GCK AQFDSAAGLYEGNTVAVLGMPVGKVVRIEPKGGYVEVEFTVDGDVAVPAD
Mvan_0962|M.vanbaalenii_PYR-1 AQFDSAAGLYEGNTVAVLGMPVGEVTKIVTKGGYVEVEFTVDGDVRVPAD
MSMEG_1146|M.smegmatis_MC2_155 AQFDSAAGLYEGNVVAVLGMPVGEVKKIRNRNGYVEVEFTVDKGVNIPAD
TH_3076|M.thermoresistible__bu AQFDSAAGLYVGNVVAVLGMPVGKVTEITPKTNYVEVTFTVDKDVKVPAD
Mb3526c|M.bovis_AF2122/97 GYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSDVKITMSVSKDVKVPVD
Rv3496c|M.tuberculosis_H37Rv GYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSDVKITMSVSKDVKVPVD
MLBr_02592|M.leprae_Br4923 VYFPAATALYTGDKVQIMGLRVGSIDSIVPAGDKMKVTFHYQNKYKVPAN
* * .** .: * ::*: ** : * . .: : . :*.:
MAV_2385|M.avium_104 AQAVTVSTSILTDRQIELTPPYRG-GPVLGNHDTIGLPRTKTPVEFSSVL
MMAR_3867|M.marinum_M AQAVTVSTSILTDRQIELTPPYRG-GPVLQSHDTIGLTRTKTPVEFSRVL
MAB_1007c|M.abscessus_ATCC_199 VQAVTLSTSILTDRQVELTPPYRG-GPTLQDGDTIGLDKTKTPVEFDRVL
MUL_4916|M.ulcerans_Agy99 VQAVTISTSILTDRQIELTPAYRG-GPTLRNHDTIGLTRTRAPVEFDRVL
Mflv_0007|M.gilvum_PYR-GCK VQAVTISNSILTDRQIELTPPYRGDGPTLRDHDTIGLNRTRTPVEFARVL
Mvan_0962|M.vanbaalenii_PYR-1 AMAVTISNSILTDRQIELTPPYRGDGPMLQNHDTIGLNRTRTPVEFARVL
MSMEG_1146|M.smegmatis_MC2_155 VQAATISNSILTDRQIELTPPYRG-GPTLQNNATIGLNRTRTPVEFARVL
TH_3076|M.thermoresistible__bu VQAVTLSNSILTDRQIELTPPYRG-GPTLQDHDTIGLNRTRTPVEFARVL
Mb3526c|M.bovis_AF2122/97 VQAVIMSPNLVAARFIQLTPVYTG-GAVLPDNGRIDLDRTAVPVEWDEVK
Rv3496c|M.tuberculosis_H37Rv VQAVIMSPNLVAARFIQLTPVYTG-GAVLPDNGRIDLDRTAVPVEWDEVK
MLBr_02592|M.leprae_Br4923 ASAVILNPTLVASRSIQLEPPYKG-GPALGDNAVIPLERTQVPVEWDELR
. *. :. .::: * ::* * * * *. * . * * :* .***: :
MAV_2385|M.avium_104 NVLDKVTKSLE-GDGHGGGPIADVLGGGAEVVNGNGEKIKAALGELSKAL
MMAR_3867|M.marinum_M SVLDKVTKSLQ-GDGHGGGPVADVLSGGTEIVDGNGAKIKGALDALSRAL
MAB_1007c|M.abscessus_ATCC_199 GMLDKLSVSLK-GDGNGQGPVSDIINAAAGVADGNGDKIKSGLDELSKAL
MUL_4916|M.ulcerans_Agy99 GMLDRLASSLQ-VDGRGGGPVAGVLDAGYDVVNGNGQHLKEAFDELSKAL
Mflv_0007|M.gilvum_PYR-GCK DVLDKLSTSLR-GDGRGGGPVADVVNASAAIADGNGQQMKDALGELSTAL
Mvan_0962|M.vanbaalenii_PYR-1 DVLDKLSTSLR-GDGKGNGPVRDVVDASAAIADGNGQAMKDALGELSNAL
MSMEG_1146|M.smegmatis_MC2_155 DVLDKLAGSLR-GDGKGNGPIADVLGAGASIVDGNGQQMKDALGELSNAL
TH_3076|M.thermoresistible__bu DVLDKLALSLR-GDGKGGGPIADVVDAGAAIVDGQGRQMKDALGELSNAL
Mb3526c|M.bovis_AF2122/97 EGLTRLAADLSPAAGELQGPLGAAINQAADTLDGNGDSLHNALRELAQVA
Rv3496c|M.tuberculosis_H37Rv EGLTRLAADLSPAAGELQGPLGAAINQAADTLDGNGDSLHNALRELAQVA
MLBr_02592|M.leprae_Br4923 DSITKIISKLGPTKEQPKGPFGDVIESFADGLAGKGKQINATLDSLSRAL
: :: .* . **. : *:* :: : *: .
MAV_2385|M.avium_104 RLSSDGGAATREQITTIVKNLSTLFDAVASNDTKLREFASTIHQVSQIMA
MMAR_3867|M.marinum_M RLSSDGGAQTREQITTIIRNVSSLFDAAAANDAKLREFASTIHQVSQILA
MAB_1007c|M.abscessus_ATCC_199 RLSSEGGVTTREQMTTIIKNVSSLFDAAAANDGKLREFSTTIHQLSNIID
MUL_4916|M.ulcerans_Agy99 RLSADRGVHTREQMTAIIENLSSLLDAAAKNDSTLRQFGSTVRQLSQIMA
Mflv_0007|M.gilvum_PYR-GCK RLSADHGQVTRDQLTTIIRNVSTLSQAAADNDATLREFGSSVRQLSQVLA
Mvan_0962|M.vanbaalenii_PYR-1 RLSSDRGEVTRDQLTTIIRNVSTLFEAAANNDATLREFGSTIRALSQILA
MSMEG_1146|M.smegmatis_MC2_155 RLSSDRGQVTKDQLTTIVRNLSALSQAAADNDAMMREFGSSVRALSQIVA
TH_3076|M.thermoresistible__bu RLSADRGELTRDQLTTIVRNVSSLAEAAARNDELLREFGSSVRAMSQILA
Mb3526c|M.bovis_AF2122/97 GRLGDS----RGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASVSQVLA
Rv3496c|M.tuberculosis_H37Rv GRLGDS----RGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASVSQVLA
MLBr_02592|M.leprae_Br4923 TALNEG----RGDFFAVLRSLALFVNALHTDDQQFVALNQNLADFTTRLA
: : :: :..: : :* .* : : : .: :
MAV_2385|M.avium_104 DEDLGSGNTGHKLDQLIQRAGDLLDANRDNIKHAVLSGNDTLK-TVTDQR
MMAR_3867|M.marinum_M DEDIGGGNTGKRLDQLVQRAGDLLDANREAIRQTVLNGNTTLQ-TVSDQQ
MAB_1007c|M.abscessus_ATCC_199 DEALGTGNTGRQLNDLVTQAGDLLDANRDHIKQSILNGNTALK-TVVDNQ
MUL_4916|M.ulcerans_Agy99 NEGFGSGTSGQQLNALIQRTGDILNANREAIARGIANGNTTIQ-SVVDRQ
Mflv_0007|M.gilvum_PYR-GCK DENFGTGTTGKKLNEVIVQFGEVLDTHREAIRQIVTNGNTAVT-TTVDHQ
Mvan_0962|M.vanbaalenii_PYR-1 DENFGSGSTGRKLNDVINQLGEVLHTHRDAIRQIVSNGDTALT-TTVEHQ
MSMEG_1146|M.smegmatis_MC2_155 DEDLGSGTTGKKINEVLTHTGQVLETHRDAIKDLVANGDVVLNTVAVDHE
TH_3076|M.thermoresistible__bu DEKLGSGTTGRKINEVLTHAGEVLQTHRETIRDLIANGDVVLN-AAVDHE
Mb3526c|M.bovis_AF2122/97 DSSANLDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQ-TLSDQS
Rv3496c|M.tuberculosis_H37Rv DSSANLDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQ-TLSDQS
MLBr_02592|M.leprae_Br4923 HSDTDLSSALQQFNSMLSTVQSFLVKNREVLAADVNNLATVNN-TVVQPE
.. . . : :: : .* :.. : : . :
MAV_2385|M.avium_104 --RDLAELLDLAPLVADNAYNMIDRANGSVRARFLTD-------------
MMAR_3867|M.marinum_M --RDLAEILDLAPLVADNGYNAIDRANGAVRAHFLTD-------------
MAB_1007c|M.abscessus_ATCC_199 --RELAETIDVAPLAVENLYNTVDQDNGSFRARFLTD-------------
MUL_4916|M.ulcerans_Agy99 --REVTEFFDVLPLMADNLYNAVDRRNGSIRAHQLTD-------------
Mflv_0007|M.gilvum_PYR-GCK --RDLAEFLDVTPMTLDNIYNAIDRTNGAVRVHVLTD-------------
Mvan_0962|M.vanbaalenii_PYR-1 --RDLAEFLDLAPLTLDNIYNAIDNKNGAVRVHVLTD-------------
MSMEG_1146|M.smegmatis_MC2_155 --RDVKELLDIAPMTLDNLYNVADKKNGALRVKVVTD-------------
TH_3076|M.thermoresistible__bu --RDLKELLDVTPMTLDNLYNVADQRNGALRIKVVTD-------------
Mb3526c|M.bovis_AF2122/97 --ENIEQVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFICGGS
Rv3496c|M.tuberculosis_H37Rv --ENIEQVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFICGGS
MLBr_02592|M.leprae_Br4923 PLNGVETFLHVAPTAAANMNQLYHPAHGSVVGVPTITNFANPMQFICS--
. : :.: * : . :*:.
MAV_2385|M.avium_104 --RLLFDSQYTKEICNLMGLRQLGCSTGTIQDFGPDFGLTYVLDGLAAMG
MMAR_3867|M.marinum_M --RMIFDSQLTKEVCNMMGLRQLGCSTGTLQDFGPDFGLTFVLDGMAAMG
MAB_1007c|M.abscessus_ATCC_199 --KLLFESQTGKEICNLMGLRQLGCSTGTLQDYGPDFGLTYVLDGLAAMG
MUL_4916|M.ulcerans_Agy99 --RMIFDSQFGKEVCNLMGLRQLGCSTGTLQDYGPDFGLTYLLDGLAAMG
Mflv_0007|M.gilvum_PYR-GCK --KVLFDTQTVKEMCNMMGLRQLGCSTGTLQDFGPDFGLSYVLDGLTAMG
Mvan_0962|M.vanbaalenii_PYR-1 --KVLFDTQTVKEMCNMMGLRQLGCSSGTLQDFGPDFGLSYVLDGLAAMG
MSMEG_1146|M.smegmatis_MC2_155 --KLLFDNQLIKEVCNMMGLRQLGCSTGTLQDFGPDFGLSSMLDGLAAMG
TH_3076|M.thermoresistible__bu --KVLFDNQLVKEVCNMMGLRQLGCSTGTLQDFGPDFGLTYMLEGLAAMG
Mb3526c|M.bovis_AF2122/97 FDTAAGPSAPDYYRRAEICRERLGPVLRRLTVNYPPIMFHPLNTITAYKG
Rv3496c|M.tuberculosis_H37Rv FDTAAGPSAPDYYRRAEICRERLGPVLRRLTVNYPPIMFHPLNTITAYKG
MLBr_02592|M.leprae_Br4923 --AIQAGSRLGYQESAELCAQYLSPVLDAIKFNYFPFGLNLFSTAEVLPK
. : . *. : : : . .
MAV_2385|M.avium_104 QK------------------------------------------------
MMAR_3867|M.marinum_M QK------------------------------------------------
MAB_1007c|M.abscessus_ATCC_199 QK------------------------------------------------
MUL_4916|M.ulcerans_Agy99 QK------------------------------------------------
Mflv_0007|M.gilvum_PYR-GCK QK------------------------------------------------
Mvan_0962|M.vanbaalenii_PYR-1 QK------------------------------------------------
MSMEG_1146|M.smegmatis_MC2_155 QK------------------------------------------------
TH_3076|M.thermoresistible__bu QR------------------------------------------------
Mb3526c|M.bovis_AF2122/97 QIIYDTPATEAKS---ETPVP--------------ELTWVPAGGG-----
Rv3496c|M.tuberculosis_H37Rv QIIYDTPATEAKS---ETPVP--------------ELTWVPAGGG-----
MLBr_02592|M.leprae_Br4923 EVAYSEARLQPPSGYKDTTVPGIWVPDTPLSHRNTQPGWIVAPGMQGQQV
:
MAV_2385|M.avium_104 --------------------------------------------------
MMAR_3867|M.marinum_M --------------------------------------------------
MAB_1007c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_4916|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0007|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0962|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1146|M.smegmatis_MC2_155 --------------------------------------------------
TH_3076|M.thermoresistible__bu --------------------------------------------------
Mb3526c|M.bovis_AF2122/97 ------------------------------APVGNPADLQSLLVPP----
Rv3496c|M.tuberculosis_H37Rv ------------------------------APVGNPADLQSLLVPP----
MLBr_02592|M.leprae_Br4923 GPITAALMTPDSLAELMGGPDIEPIQSNLQTPPGPPTAYDEHPVPPPIGL
MAV_2385|M.avium_104 -----------------------------------------
MMAR_3867|M.marinum_M -----------------------------------------
MAB_1007c|M.abscessus_ATCC_199 -----------------------------------------
MUL_4916|M.ulcerans_Agy99 -----------------------------------------
Mflv_0007|M.gilvum_PYR-GCK -----------------------------------------
Mvan_0962|M.vanbaalenii_PYR-1 -----------------------------------------
MSMEG_1146|M.smegmatis_MC2_155 -----------------------------------------
TH_3076|M.thermoresistible__bu -----------------------------------------
Mb3526c|M.bovis_AF2122/97 -------------APGPAPAPPAPGAGPG------EHGGGG
Rv3496c|M.tuberculosis_H37Rv -------------APGPAPAPPAPGAGPG------EHGGGG
MLBr_02592|M.leprae_Br4923 QASVPIPSPRPDVVPGPVAPTPSPAANVGDPPLPAELGAGQ