For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RTLVLGGIRSGKSQWAEEAVADGLPAGRRVRYLAPGPPGVHDAAWAHRVAEHRDRRPAHWSTIETDDIAT ELRRCPQVPTLVDDLGGWLTGMLDRHDGWQGGSVTAPVDDLLAAVAEFAAALVLVSPEVGLTVVPATASA RRFADELGSLNQRLARLCDRVVLVVAGQPVPIKTPGV*
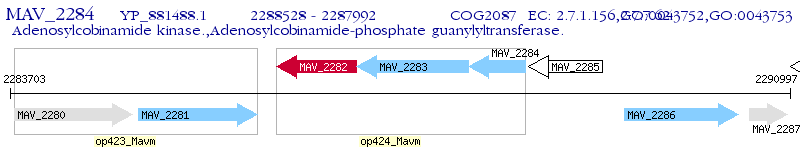
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2284 | cobU | - | 100% (178) | cobinamide kinase / cobinamide phosphate guanyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0260c | cobU | 5e-30 | 44.89% (176) | putative bifunctional cobalamin biosynthesis protein cobU: |
| M. gilvum PYR-GCK | Mflv_0355 | - | 2e-31 | 48.00% (175) | cobalbumin biosynthesis enzyme |
| M. tuberculosis H37Rv | Rv0254c | cobU | 5e-30 | 44.89% (176) | bifunctional cobinamide kinase/cobinamide phosphate |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1954c | - | 9e-52 | 59.77% (174) | putative cobinamide kinase/cobinamide phosphate |
| M. marinum M | MMAR_3251 | cobU | 4e-66 | 68.68% (182) | bifunctional cobalamin biosynthesis protein CobU |
| M. smegmatis MC2 155 | MSMEG_4274 | cobU | 5e-57 | 63.43% (175) | cobinamide kinase / cobinamide phosphate guanyltransferase |
| M. thermoresistible (build 8) | TH_1435 | cobU | 6e-64 | 66.48% (176) | Cobinamide kinase / cobinamide phosphate guanyltransferase |
| M. ulcerans Agy99 | MUL_3564 | cobU | 8e-67 | 69.23% (182) | bifunctional cobalamin biosynthesis protein CobU |
| M. vanbaalenii PYR-1 | Mvan_0379 | - | 5e-34 | 48.02% (177) | cobalbumin biosynthesis enzyme |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3251|M.marinum_M -MRTLVLGGIRSGKSRWAEETIADSLTPE--QPVRYLASGPATDS-DSAA
MUL_3564|M.ulcerans_Agy99 -MRTLVLGGIRSGKSRWAEETIADSLTPE--QPVRYLASGPATDS-DSAA
MAV_2284|M.avium_104 -MRTLVLGGIRSGKSQWAEEAVADGLPAG--RRVRYLAPGPPGVH-DAA-
MSMEG_4274|M.smegmatis_MC2_155 ---MLVLGGIRSGKSQWAESVAQQLAGEGDRAPVRYVATGPVPEG-DAD-
TH_1435|M.thermoresistible__bu -VRILVLGGIRSGKSQWAETALADAVGD---APARYLATGRVPDA-DTD-
MAB_1954c|M.abscessus_ATCC_199 MATTLVLGGIRSGKSRFAESLLP---AEG---PARYLATGSPDTG-DPA-
Mb0260c|M.bovis_AF2122/97 -MRILVTGGVRSGKSTHAEALLGDA------ADVVYVAPGRPAAGSDPD-
Rv0254c|M.tuberculosis_H37Rv -MRILVTGGVRSGKSTHAEALLGDA------ADVVYVAPGRPAAGSDPD-
Mflv_0355|M.gilvum_PYR-GCK -MRILVTGGVRSGKSRHAEELLAGA------GHVTYVAPGRAADGTDPD-
Mvan_0379|M.vanbaalenii_PYR-1 -MMILVTGGVRSGKSRHAEALLSDA------RQVTYLAPGRPADGSDPD-
** **:***** ** . *:*.* *.
MMAR_3251|M.marinum_M SQDWDQRVARHRDRRPGHWSTVETDDITTQLRQVPDTPTLVDDLGTWLTS
MUL_3564|M.ulcerans_Agy99 SQDWDQRVARHRDRRPGHWSTVETDDIATQLRQVPDTPTLVDDLGTWLTS
MAV_2284|M.avium_104 ---WAHRVAEHRDRRPAHWSTIETDDIATELRRCPQVPTLVDDLGGWLTG
MSMEG_4274|M.smegmatis_MC2_155 ---WATRVEAHQVRRPAHWQTVETTDLTATLTAAPHVATLVDDIGGWLTA
TH_1435|M.thermoresistible__bu ---WAARVSAHRRRRPAHWSTCETTDVATQLRTYPDLPTMIDDVGGWLTA
MAB_1954c|M.abscessus_ATCC_199 ---WTERIAAHRSRRPSQWTTAETTDIATELRTRAPLPTLVDDLGGWLTA
Mb0260c|M.bovis_AF2122/97 ---WDARVALHRARRPPTWLTVETADVATALSEARS-PVLVDCLGTWLTA
Rv0254c|M.tuberculosis_H37Rv ---WDARVALHRARRPPTWLTVETADVATALSEARS-PVLVDCLGTWLTA
Mflv_0355|M.gilvum_PYR-GCK ---WDARVSRHRARRPAHWRTVETADVAVAVRDAQG-TVLVDCLGTWLTA
Mvan_0379|M.vanbaalenii_PYR-1 ---WDARVAGHRTRRPAHWQTVETADVAAAVRDADG-AVLVDCLGTWLTA
* *: *: *** * * ** *::. : ..::* :* ***.
MMAR_3251|M.marinum_M AMDSHRAWEGGSP--PAALQDTIDDMLAAVGEFGSTLVLVSPEVGLTVVP
MUL_3564|M.ulcerans_Agy99 AMDSHRAWEGGSP--PAALQDTIDDMLAAVGEFGSTLVLVSPEVGLTVVP
MAV_2284|M.avium_104 MLDRHDGWQGGS------VTAPVDDLLAAVAEFAAALVLVSPEVGLTVVP
MSMEG_4274|M.smegmatis_MC2_155 TMDRRDAWAGHP------VTGDVTALVDAVAAFRAPLVMVSPEVGLTVVP
TH_1435|M.thermoresistible__bu TMDRHDAWSGGS------VTADTDELVAAVADFRSTLLVVSPEVGLSVVP
MAB_1954c|M.abscessus_ATCC_199 AMDAHGAWDVGAG----AVSADIGALVEAVADYPVDLVIVSPEVGLTVVP
Mb0260c|M.bovis_AF2122/97 IMDGEALWSAATADVYAVLEARLDGLCAALTG-LPTAIVVTNEVGLGVVP
Rv0254c|M.tuberculosis_H37Rv IMDGEALWSAATADVYAVLEARLDGLCAALTG-LPTAIVVTNEVGLGVVP
Mflv_0355|M.gilvum_PYR-GCK VLDDNGLWDKGSREATDEVEIRVTALCDALAG-AAGAVVVTNEVGLGVVP
Mvan_0379|M.vanbaalenii_PYR-1 VLDRNNLWDKGSREVHDAVDARLTALCAALTARREAVVVVTNEVGLGVVP
:* . * . : : *: ::*: **** ***
MMAR_3251|M.marinum_M ATASGRRFADELGTLNQRMAALCDRVVLVVAGQAVPIKPSRA
MUL_3564|M.ulcerans_Agy99 ATASGRRFADELGTLNQRMAALCDRVVLVVAGQAVPIKPSRA
MAV_2284|M.avium_104 ATASARRFADELGSLNQRLARLCDRVVLVVAGQPVPIKTPGV
MSMEG_4274|M.smegmatis_MC2_155 ATAAGRRFADELGTVNQRLAAVCDRVVLVVAGQPVTVKEPT-
TH_1435|M.thermoresistible__bu TTAVGRRFADALGTLNQRLARVCDRVVLVVAGQPVTVKEPV-
MAB_1954c|M.abscessus_ATCC_199 ATAAGRLFADALGALNQAVAARADRVLLVVAGQPLTIKGGRS
Mb0260c|M.bovis_AF2122/97 SHSSGVLFRDLLGTINRRVAAVCDEVHLVIAGRVLKL-----
Rv0254c|M.tuberculosis_H37Rv SHSSGVLFRDLLGTINRRVAAVCDEVHLVIAGRVLKL-----
Mflv_0355|M.gilvum_PYR-GCK SHRSGVLFRDLLGTVNQRVAQVCDEVHLVIAGRVLTL-----
Mvan_0379|M.vanbaalenii_PYR-1 SHRSGVLFRDLLGTANQRVAAVCDEVHLVIAGRVLKI-----
: . * * **: *: :* .*.* **:**: : :