For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSVSRLQQLCGIGVNDVGDRADALNDPEMLRLENLDTDLAPPAVAVEVTKRAVTDDAANSYLPFEGHYEL RRAAATHVSRISGISYDPFRECVSVAGGTNGVLNALLATVEPGKEVVIADPTYAGLINRIRLAGAIPRHI AAHPSPTGWRTDPAELAAAIGPNTAAVLLMGPEMPTGALLSSEHLDAITEPVTQHRAWVIYDAAMERIRF DDQPPLHPASHPGLAERTITIGSASKELRMIGWRVGWIVGPRAIMADIALVGLTNVVCQGGIVKTCGLGC FRRPPFRLIRPVRVV
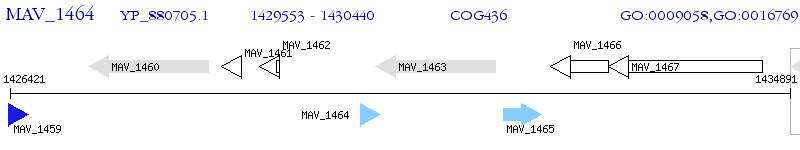
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1464 | - | - | 100% (295) | aminotransferase, classes I and II family protein |
| M. avium 104 | MAV_0979 | - | 9e-32 | 35.48% (217) | aminotransferase |
| M. avium 104 | MAV_4818 | - | 2e-15 | 28.28% (198) | aminotransferase AlaT |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0881c | - | 1e-31 | 37.50% (200) | aminotransferase |
| M. gilvum PYR-GCK | Mflv_1680 | - | 9e-33 | 34.86% (218) | aminotransferase |
| M. tuberculosis H37Rv | Rv0858c | - | 1e-31 | 37.50% (200) | aminotransferase |
| M. leprae Br4923 | MLBr_02502 | - | 2e-16 | 28.79% (198) | aminotransferase AlaT |
| M. abscessus ATCC 19977 | MAB_0847c | - | 3e-32 | 34.56% (217) | aminotransferase |
| M. marinum M | MMAR_4678 | - | 3e-28 | 34.39% (221) | aminotransferase |
| M. smegmatis MC2 155 | MSMEG_5725 | - | 3e-34 | 35.48% (217) | aminotransferase |
| M. thermoresistible (build 8) | TH_2032 | - | 5e-29 | 34.82% (224) | PLP-dependent aminotransferases |
| M. ulcerans Agy99 | MUL_0297 | - | 3e-27 | 33.94% (221) | aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_5070 | - | 9e-29 | 33.94% (218) | aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0881c|M.bovis_AF2122/97 ------------------------------------MTVSRLRPYATTVF
Rv0858c|M.tuberculosis_H37Rv ------------------------------------MTVSRLRPYATTVF
MMAR_4678|M.marinum_M ------------------------------------MTVSRLQPYATTVF
MUL_0297|M.ulcerans_Agy99 ------------------------------------MTVSRLQPYATTVF
TH_2032|M.thermoresistible__bu ------------------------------------MTVERLRPYATTIF
Mflv_1680|M.gilvum_PYR-GCK ------------------------------------MTVQRLRPYAVTIF
Mvan_5070|M.vanbaalenii_PYR-1 ------------------------------------MTVRRLQPYAVTIF
MSMEG_5725|M.smegmatis_MC2_155 ------------------------------------MTVRRLQPFAVTIF
MAB_0847c|M.abscessus_ATCC_199 ---------------------------------MNQRTVERLRPFGATIF
MAV_1464|M.avium_104 ------------------------------------MSVSRLQQLCGIGV
MLBr_02502|M.leprae_Br4923 MTLYQQISVDGSGTMSDVTTHQLPMHTPGYHRQRAFAQSSKLQDVLYEIR
:*:
Mb0881c|M.bovis_AF2122/97 AEMSALATRIG-------AVNLG-QGFPDEDGPPKMLQAAQDAIAGGVN-
Rv0858c|M.tuberculosis_H37Rv AEMSALATRIG-------AVNLG-QGFPDEDGPPKMLQAAQDAIAGGVN-
MMAR_4678|M.marinum_M AEMSALAARIG-------AVNLG-QGFPDEDGPPAMLKAAQEAIAAGAN-
MUL_0297|M.ulcerans_Agy99 AEMSALAARIG-------AVNLG-QGFPDEDGPPAMLKAAQEAIAAGAN-
TH_2032|M.thermoresistible__bu AEMSALAARVG-------AVNLG-QGFPDEDGPPAMLEIARDAIAEGMN-
Mflv_1680|M.gilvum_PYR-GCK AETSALAARVG-------AVNLG-QGFPDEDGPPAMLKAAENAIAEGVN-
Mvan_5070|M.vanbaalenii_PYR-1 AEMSALAARVG-------AVNLG-QGFPDEDGPPAMLKAAENAIAEGVN-
MSMEG_5725|M.smegmatis_MC2_155 AEMSALATRLG-------AVNLG-QGFPDEDGPVEMLKVAQNAIADGIN-
MAB_0847c|M.abscessus_ATCC_199 AEMSALAVRHD-------AINLG-QGFPDEDGPASMLDAAQQAIRSGLN-
MAV_1464|M.avium_104 NDVGDRADALNDP----EMLRLE-NLDTDLAPPAVAVEVTKRAVTDDAAN
MLBr_02502|M.leprae_Br4923 GPVSQHATRLETEGHRILKLNIGNPALFGFDAPDVIMRDMIQALPYAQG-
. * :.: . * : *:
Mb0881c|M.bovis_AF2122/97 QYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGATEAIAAAVLGL
Rv0858c|M.tuberculosis_H37Rv QYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGATEAIAAAVLGL
MMAR_4678|M.marinum_M QYPPGMGIPALRQAIAAQRRRRFGVEYDPDTEVLVTVGATEAIAAAVLGL
MUL_0297|M.ulcerans_Agy99 QYPPGMGIPALRQAIAAQRRRRFGVEYDPDTEVLVTVGATEAIAAAVRGL
TH_2032|M.thermoresistible__bu QYPPGIGIAPLRQAIAAQRERHYGVEYDPDTEVLVTVGATEAIAGAVLGL
Mflv_1680|M.gilvum_PYR-GCK QYPPGLGFAPLRQAIADQRRRRYGTVYDPDTEVLVTVGATEAIASSVLGL
Mvan_5070|M.vanbaalenii_PYR-1 QYPPGLGIAPLRQAIADQRRRRYGSEYDPDTEVLVTVGATEAIAAAILGL
MSMEG_5725|M.smegmatis_MC2_155 QYPPGLGVPELRQAIAAQRKRVHGVQYDPDTEVLVTVGATEAIAASVIGL
MAB_0847c|M.abscessus_ATCC_199 QYPPGLGIPELRRAITADRLARYGEELDPDTQVLVTVGATEAIAGALLGL
MAV_1464|M.avium_104 SYLPFEGHYELRRAAATHVSRISGISYDPFRECVSVAGGTNGVLNALLAT
MLBr_02502|M.leprae_Br4923 -YSDSQGILPARRAVVTRYELVDGFPRFDVDDVYLGNGVSELITMTLQAL
* * *:* . * : * :: : :: .
Mb0881c|M.bovis_AF2122/97 VEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRGFALDADALRRA
Rv0858c|M.tuberculosis_H37Rv VEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRGFALDADALRRA
MMAR_4678|M.marinum_M VEPGSEVLLIEPFYDSYAPVIAMAGAKRVAVPLVPDGRGFALDADALRRA
MUL_0297|M.ulcerans_Agy99 VEPGSEVLLIEPFYDSYAPVIAMAGAKRVAVPLVPDGRGFALDADALRRA
TH_2032|M.thermoresistible__bu VEPGSEVLLIEPFYDSYSPVIAMAGAHRVAVPLVTDGPGFAVDLDALRRA
Mflv_1680|M.gilvum_PYR-GCK VEPGSEVLLIEPFYDSYSPVIAMAGCHRRAVPLRQNGRGFAIDIEALRAA
Mvan_5070|M.vanbaalenii_PYR-1 VEPDSEVLLIEPFYDSYSPVIAMAGCHRRAVPMVRDGRGFAIDVEGLRRA
MSMEG_5725|M.smegmatis_MC2_155 VEPGSEVLVIEPFYDSYSPVIAMAGCERRAVPLVPSGRGFAIDVDALRAA
MAB_0847c|M.abscessus_ATCC_199 VEPGSEVILIEPYYDSYAAVVAMAGAVRVPVPLVPDGAGFALDTDALAAA
MAV_1464|M.avium_104 VEPGKEVVIADPTYAGLINRIRLAGAIPRHIAAHPSPTGWRTDPAELAAA
MLBr_02502|M.leprae_Br4923 LDNGDQVLIPSPDYPLWTASTSLAGGTPVHY-LCDETQGWQPDIADVESK
:: ..:*:: .* * :** . *: * :
Mb0881c|M.bovis_AF2122/97 VTPRTRALIINSPHNPTGAVLSATELAAIAEIAVAANLVVITDEVYEHLV
Rv0858c|M.tuberculosis_H37Rv VTPRTRALIINSPHNPTGAVLSATELAAIAEIAVAANLVVITDEVYEHLV
MMAR_4678|M.marinum_M VTPKTRALIVNSPHNPTGAVLRRAELAAIAEIAVAADLLVITDEVYEHLV
MUL_0297|M.ulcerans_Agy99 VTPKTRALIVNSPHNPTGAVLSRAELAAIAEIAVAADLLVITDEVYEHLV
TH_2032|M.thermoresistible__bu VTPRTRAMIVNSPHNPTGMVLTEAELAGIAELAVSADLLVITDEVYEHLV
Mflv_1680|M.gilvum_PYR-GCK VGPRTRALIVNTPHNPTGMIASDEELRGLAELAVAHDLLVITDEVYEHLV
Mvan_5070|M.vanbaalenii_PYR-1 VTPKTRALILNSPHNPTGSVASDDELRAVAELAISADLLVITDEVYEHLV
MSMEG_5725|M.smegmatis_MC2_155 VTPQTKALILNSPHNPTGAVASDAELRAVAALAVEHDLLVITDEVYEALV
MAB_0847c|M.abscessus_ATCC_199 ITPRTAALVINSPHNPTGKVFTDTELARIAELAVEHDLLVVSDEVYERLL
MAV_1464|M.avium_104 IGPNTAAVLLMGPEMPTGALLSSEHLDAITEPVTQHRAWVIYDAAMERIR
MLBr_02502|M.leprae_Br4923 ITERTKALVVINPNNPTGAVYSNEILNQIVDLARKHQLLLLADEIYDKIL
: .* *::: *. *** : * :. . :: * : :
Mb0881c|M.bovis_AF2122/97 FDHAR---HLPLAGFDGMAERTITISSAAKMFNCTGWKIGWACGP-----
Rv0858c|M.tuberculosis_H37Rv FDHAR---HLPLAGFDGMAERTITISSAAKMFNCTGWKIGWACGP-----
MMAR_4678|M.marinum_M FTSPTGHQHLPLAGFDGMAQRTVTISSAAKMFNCTGWKIGWACGP-----
MUL_0297|M.ulcerans_Agy99 FTSPTGHQHLPLAGFDGMAQRTVTISSAAKMFNCTGWKIGWACGL-----
TH_2032|M.thermoresistible__bu YPSPAGRRHIPLASYPGMAERTVTISSAAKMFNCTGWKIGWACGP-----
Mflv_1680|M.gilvum_PYR-GCK FDGRQ---HRPLAGYPGMAERTVTISSAGKMFNVTGWKIGWACGP-----
Mvan_5070|M.vanbaalenii_PYR-1 FDGRR---HRPLADYPGMAGRTITISSAGKMFNVTGWKIGWACGP-----
MSMEG_5725|M.smegmatis_MC2_155 FDGHR---HLTLASYPGMRDRTVTISSAAKMFNATGWKIGWACGA-----
MAB_0847c|M.abscessus_ATCC_199 FDGRT---QTPMASLPGMADRTVTISSAAKTFNCTGWKIGWACGT-----
MAV_1464|M.avium_104 FDDQP---PLHPASHPGLAERTITIGSASKELRMIGWRVGWIVGP-----
MLBr_02502|M.leprae_Br4923 YDDTK---HISVASIA-PDLLCLTFNGLSKAYRVAGYRSGWLAITGPKDH
: *. :*:.. .* . *:: **
Mb0881c|M.bovis_AF2122/97 -AELIAGVRAAKQYLSYVGGAPFQPAVALALDTEDAWVAALRNSLRARRD
Rv0858c|M.tuberculosis_H37Rv -AELIAGVRAAKQYLSYVGGAPFQPAVALALDTEDAWVAALRNSLRARRD
MMAR_4678|M.marinum_M -QELIAGVRAAKQYMSYVGGAPLQPAVALALDSEDAWVAALRTSLQARRD
MUL_0297|M.ulcerans_Agy99 -KELIAGVRAAKQYMSYVGGAPLQPAVALALDSEDAWVVALRTSLQARRD
TH_2032|M.thermoresistible__bu -SDLIAGVRAAKQYLSYVGGAPFQPAVAHALDHEDAWVAALRDSLQSRRD
Mflv_1680|M.gilvum_PYR-GCK -SDLIAGVRAAKQYLSYVAGAPFQPAVAQALNNEDAWVDALCASFQARRD
Mvan_5070|M.vanbaalenii_PYR-1 -SDLIAGVRAAKQYLSYVGGAPFQPAVAHALATEDAWVDALCASFQSRRD
MSMEG_5725|M.smegmatis_MC2_155 -PELIAGVRAAKQYLTYVGGAPFQPAVAHALNHEDGWVADLRDSFQSKRD
MAB_0847c|M.abscessus_ATCC_199 -PELVAGVRAAKQFLSYVGGGPFQPAVAVALDTEQSWVKTLRESLQRRRD
MAV_1464|M.avium_104 -RAIMADIALVGLTNVVCQGG-----------------------------
MLBr_02502|M.leprae_Br4923 ASSFIGGINLLAN-MRLCPNVPAQHAIQVALGGHQSIEDLVLPGGRLLEQ
::..: .
Mb0881c|M.bovis_AF2122/97 R-----LAAGLTEIGFAVHDSYGTYFLCADPRPLGYDDSTEFCAALPEKV
Rv0858c|M.tuberculosis_H37Rv R-----LAAGLTEIGFAVHDSYGTYFLCADPRPLGYDDSTEFCAALPEKV
MMAR_4678|M.marinum_M R-----LAAGLSEIGFGVHDSHGTYFLCADPRPLGYDDSTAFCAALPEKV
MUL_0297|M.ulcerans_Agy99 R-----LAAGLSEIGFGVHDSHGTYFLCADPRPLGYDDSTAFCAALPERV
TH_2032|M.thermoresistible__bu T-----LAAALTEIGFEVHDSAGTYFLCADPRPLGYPDSTEFCAGLPERV
Mflv_1680|M.gilvum_PYR-GCK R-----LSSALSDIGFDVFDSFGTYFLTVDPRPLGYDDSTSFCAQLPEQV
Mvan_5070|M.vanbaalenii_PYR-1 R-----LGSALSDIGFEVHESFGTYFLCVDPRPLGFTDSAAFCAELPERV
MSMEG_5725|M.smegmatis_MC2_155 R-----LGSALAEIGFEVHDSRGTYFLCADPRPLGYDDSTAFCAELPTRA
MAB_0847c|M.abscessus_ATCC_199 R-----LSAALTGLGFEVHSSDGGYFVCADPRPLGFSDSAELCRRLPETV
MAV_1464|M.avium_104 ---------------------------IVKTCGLGCFRRPPFRLIRPVRV
MLBr_02502|M.leprae_Br4923 RDVAWTKLNEIPGVSCVKPTGALYAFPRLDPEVYGIVDDEQLVLDLLLQE
.. * :
Mb0881c|M.bovis_AF2122/97 GVAAIPMSAFCDPAAGQASQQADVWNHLVRFTFCKRDDTLDEAIRRLSVL
Rv0858c|M.tuberculosis_H37Rv GVAAIPMSAFCDPAAGQASQQADVWNHLVRFTFCKRDDTLDEAIRRLSVL
MMAR_4678|M.marinum_M GVAAIPMSAFCDPVAEHASTQADIWNHLVRFTFCKREDTLDEAIKRLAAL
MUL_0297|M.ulcerans_Agy99 GVAAIPMSAFCDPVAEHAPTQADIWNHLVRFTFCKREDTLDEAIKRLAAL
TH_2032|M.thermoresistible__bu GVAAIPMSAFCDPAAPHA----EVWNHLVRFTFCKRPETLEEGIRRLGAL
Mflv_1680|M.gilvum_PYR-GCK GVAAIPMSAFCDPDAAHA----GEWKHLVRFAFCKRDETLDEAIRRLQAL
Mvan_5070|M.vanbaalenii_PYR-1 GVAAIPMSAFCDPAAGHA----EEWKHLVRFAFCKREDTLDEAIRRLRAL
MSMEG_5725|M.smegmatis_MC2_155 GVAAIPMSAFCDPDAPHA----DVWNHLVRFAFCKRDDTLDEAIRRLQAL
MAB_0847c|M.abscessus_ATCC_199 GVAAVPVSAFCD---RYA----DQWNHLVRFTFAKRDVVIDEAITRLARL
MAV_1464|M.avium_104 V-------------------------------------------------
MLBr_02502|M.leprae_Br4923 KILVTQGSGFNWPAPDHLR--------LVTLPWSRDLAAAIDRLGNFLVS
Mb0881c|M.bovis_AF2122/97 AERPAT
Rv0858c|M.tuberculosis_H37Rv AERPAT
MMAR_4678|M.marinum_M RDAA--
MUL_0297|M.ulcerans_Agy99 RDAA--
TH_2032|M.thermoresistible__bu RRG---
Mflv_1680|M.gilvum_PYR-GCK RSTG--
Mvan_5070|M.vanbaalenii_PYR-1 PRSR--
MSMEG_5725|M.smegmatis_MC2_155 RRC---
MAB_0847c|M.abscessus_ATCC_199 GA----
MAV_1464|M.avium_104 ------
MLBr_02502|M.leprae_Br4923 YRQ---