For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVRRLQPYAVTIFAEMSALAARVGAVNLGQGFPDEDGPPAMLKAAENAIAEGVNQYPPGLGIAPLRQAI ADQRRRRYGSEYDPDTEVLVTVGATEAIAAAILGLVEPDSEVLLIEPFYDSYSPVIAMAGCHRRAVPMVR DGRGFAIDVEGLRRAVTPKTRALILNSPHNPTGSVASDDELRAVAELAISADLLVITDEVYEHLVFDGRR HRPLADYPGMAGRTITISSAGKMFNVTGWKIGWACGPSDLIAGVRAAKQYLSYVGGAPFQPAVAHALATE DAWVDALCASFQSRRDRLGSALSDIGFEVHESFGTYFLCVDPRPLGFTDSAAFCAELPERVGVAAIPMSA FCDPAAGHAEEWKHLVRFAFCKREDTLDEAIRRLRALPRSR
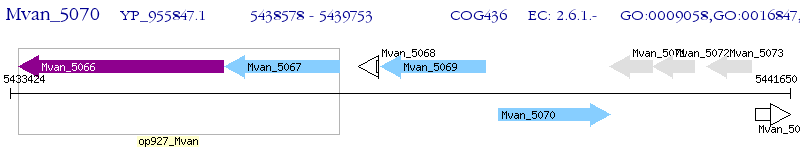
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5070 | - | - | 100% (391) | aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_5290 | - | 1e-32 | 31.40% (363) | aspartate aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_4528 | - | 1e-19 | 28.24% (386) | N-succinyldiaminopimelate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0881c | - | 1e-175 | 76.47% (391) | aminotransferase |
| M. gilvum PYR-GCK | Mflv_1680 | - | 0.0 | 85.79% (387) | aminotransferase |
| M. tuberculosis H37Rv | Rv0858c | - | 1e-175 | 76.47% (391) | aminotransferase |
| M. leprae Br4923 | MLBr_01794 | - | 6e-22 | 28.20% (383) | aminotransferase |
| M. abscessus ATCC 19977 | MAB_0847c | - | 1e-148 | 66.32% (386) | aminotransferase |
| M. marinum M | MMAR_4678 | - | 1e-175 | 77.41% (394) | aminotransferase |
| M. avium 104 | MAV_0979 | - | 1e-172 | 76.86% (389) | aminotransferase |
| M. smegmatis MC2 155 | MSMEG_5725 | - | 1e-180 | 79.69% (389) | aminotransferase |
| M. thermoresistible (build 8) | TH_2032 | - | 0.0 | 80.10% (392) | PLP-dependent aminotransferases |
| M. ulcerans Agy99 | MUL_0297 | - | 1e-174 | 77.41% (394) | aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5070|M.vanbaalenii_PYR-1 ---MTVRRLQPYAVTIFAEMSALAARVGAVNLGQGFPDEDG---PPAMLK
Mflv_1680|M.gilvum_PYR-GCK ---MTVQRLRPYAVTIFAETSALAARVGAVNLGQGFPDEDG---PPAMLK
MSMEG_5725|M.smegmatis_MC2_155 ---MTVRRLQPFAVTIFAEMSALATRLGAVNLGQGFPDEDG---PVEMLK
Mb0881c|M.bovis_AF2122/97 ---MTVSRLRPYATTVFAEMSALATRIGAVNLGQGFPDEDG---PPKMLQ
Rv0858c|M.tuberculosis_H37Rv ---MTVSRLRPYATTVFAEMSALATRIGAVNLGQGFPDEDG---PPKMLQ
MAV_0979|M.avium_104 ---MTVSRLRPYATTVFAEMSALAARVEAVNLGQGFPDEDG---PAEMLK
MMAR_4678|M.marinum_M ---MTVSRLQPYATTVFAEMSALAARIGAVNLGQGFPDEDG---PPAMLK
MUL_0297|M.ulcerans_Agy99 ---MTVSRLQPYATTVFAEMSALAARIGAVNLGQGFPDEDG---PPAMLK
TH_2032|M.thermoresistible__bu ---MTVERLRPYATTIFAEMSALAARVGAVNLGQGFPDEDG---PPAMLE
MAB_0847c|M.abscessus_ATCC_199 MNQRTVERLRPFGATIFAEMSALAVRHDAINLGQGFPDEDG---PASMLD
MLBr_01794|M.leprae_Br4923 ---MTSNPLEQLTIEQLQCRTSMKWRAHPEDVLPLWVAEMDVLLAPTVAE
* *. : ::: * . :: : * . . : .
Mvan_5070|M.vanbaalenii_PYR-1 AAENAIAEGVNQYPPGLGIAPLRQAIADQRRRRYGSEYDPDTEVLVTVGA
Mflv_1680|M.gilvum_PYR-GCK AAENAIAEGVNQYPPGLGFAPLRQAIADQRRRRYGTVYDPDTEVLVTVGA
MSMEG_5725|M.smegmatis_MC2_155 VAQNAIADGINQYPPGLGVPELRQAIAAQRKRVHGVQYDPDTEVLVTVGA
Mb0881c|M.bovis_AF2122/97 AAQDAIAGGVNQYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGA
Rv0858c|M.tuberculosis_H37Rv AAQDAIAGGVNQYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGA
MAV_0979|M.avium_104 AAQEAIADGVNQYPPGIGIAPLRHAIAAQRHRRYGIEYDPDTEVLVTVGA
MMAR_4678|M.marinum_M AAQEAIAAGANQYPPGMGIPALRQAIAAQRRRRFGVEYDPDTEVLVTVGA
MUL_0297|M.ulcerans_Agy99 AAQEAIAAGANQYPPGMGIPALRQAIAAQRRRRFGVEYDPDTEVLVTVGA
TH_2032|M.thermoresistible__bu IARDAIAEGMNQYPPGIGIAPLRQAIAAQRERHYGVEYDPDTEVLVTVGA
MAB_0847c|M.abscessus_ATCC_199 AAQQAIRSGLNQYPPGLGIPELRRAITADRLARYGEELDPDTQVLVTVGA
MLBr_01794|M.leprae_Br4923 AVHRAVDNGDTGYPYGTTFAEAISEFASQRWQWHDLDVS---RTAIVPDV
.. *: * . ** * . :: :* .. . .. :. ..
Mvan_5070|M.vanbaalenii_PYR-1 TEAIAAAILGLVEPDSEVLLIEPFYDSYSPVIAMAGCHRRAVPMVRDGRG
Mflv_1680|M.gilvum_PYR-GCK TEAIASSVLGLVEPGSEVLLIEPFYDSYSPVIAMAGCHRRAVPLRQNGRG
MSMEG_5725|M.smegmatis_MC2_155 TEAIAASVIGLVEPGSEVLVIEPFYDSYSPVIAMAGCERRAVPLVPSGRG
Mb0881c|M.bovis_AF2122/97 TEAIAAAVLGLVEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRG
Rv0858c|M.tuberculosis_H37Rv TEAIAAAVLGLVEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRG
MAV_0979|M.avium_104 TEAIASAVIGLVEPGSEVLLIEPFYDSYSPVIAMASAQRVAVPLVPHGRG
MMAR_4678|M.marinum_M TEAIAAAVLGLVEPGSEVLLIEPFYDSYAPVIAMAGAKRVAVPLVPDGRG
MUL_0297|M.ulcerans_Agy99 TEAIAAAVRGLVEPGSEVLLIEPFYDSYAPVIAMAGAKRVAVPLVPDGRG
TH_2032|M.thermoresistible__bu TEAIAGAVLGLVEPGSEVLLIEPFYDSYSPVIAMAGAHRVAVPLVTDGPG
MAB_0847c|M.abscessus_ATCC_199 TEAIAGALLGLVEPGSEVILIEPYYDSYAAVVAMAGAVRVPVPLVPDGAG
MLBr_01794|M.leprae_Br4923 MLGVVEMLRLITDRGDGVIVSPPVYPPFYAFVTHDGRRVLEAPLGRDGR-
.:. : :.: .. *:: * * .: ..:: . .*: *
Mvan_5070|M.vanbaalenii_PYR-1 FAIDVEGLRRAVTPKTR--------ALILNSPHNPTGSVASDDELRAVAE
Mflv_1680|M.gilvum_PYR-GCK FAIDIEALRAAVGPRTR--------ALIVNTPHNPTGMIASDEELRGLAE
MSMEG_5725|M.smegmatis_MC2_155 FAIDVDALRAAVTPQTK--------ALILNSPHNPTGAVASDAELRAVAA
Mb0881c|M.bovis_AF2122/97 FALDADALRRAVTPRTR--------ALIINSPHNPTGAVLSATELAAIAE
Rv0858c|M.tuberculosis_H37Rv FALDADALRRAVTPRTR--------ALIINSPHNPTGAVLSATELAAIAE
MAV_0979|M.avium_104 FALDADALRRAVTPRTR--------ALIVNSPHNPTGTVLGATELAAIAE
MMAR_4678|M.marinum_M FALDADALRRAVTPKTR--------ALIVNSPHNPTGAVLRRAELAAIAE
MUL_0297|M.ulcerans_Agy99 FALDADALRRAVTPKTR--------ALIVNSPHNPTGAVLSRAELAAIAE
TH_2032|M.thermoresistible__bu FAVDLDALRRAVTPRTR--------AMIVNSPHNPTGMVLTEAELAGIAE
MAB_0847c|M.abscessus_ATCC_199 FALDTDALAAAITPRTA--------ALVINSPHNPTGKVFTDTELARIAE
MLBr_01794|M.leprae_Br4923 --LDLAALQDAFSRARRSSGPNGKVAYLLCNPHNPTGSVHTVAELHGVAE
:* .* *. * :: .****** : ** :*
Mvan_5070|M.vanbaalenii_PYR-1 LAISADLLVITDEVYEHLVFDG---RRHRPLADYPGMAGRTITISSAGKM
Mflv_1680|M.gilvum_PYR-GCK LAVAHDLLVITDEVYEHLVFDG---RQHRPLAGYPGMAERTVTISSAGKM
MSMEG_5725|M.smegmatis_MC2_155 LAVEHDLLVITDEVYEALVFDG---HRHLTLASYPGMRDRTVTISSAAKM
Mb0881c|M.bovis_AF2122/97 IAVAANLVVITDEVYEHLVFDH---ARHLPLAGFDGMAERTITISSAAKM
Rv0858c|M.tuberculosis_H37Rv IAVAANLVVITDEVYEHLVFDH---ARHLPLAGFDGMAERTITISSAAKM
MAV_0979|M.avium_104 VAVESDLLVITDEVYEHLVFDD---HRHLPLAGFDGMAERTITISSAAKM
MMAR_4678|M.marinum_M IAVAADLLVITDEVYEHLVFTSPTGHQHLPLAGFDGMAQRTVTISSAAKM
MUL_0297|M.ulcerans_Agy99 IAVAADLLVITDEVYEHLVFTSPTGHQHLPLAGFDGMAQRTVTISSAAKM
TH_2032|M.thermoresistible__bu LAVSADLLVITDEVYEHLVYPSPAGRRHIPLASYPGMAERTVTISSAAKM
MAB_0847c|M.abscessus_ATCC_199 LAVEHDLLVVSDEVYERLLFDG---RTQTPMASLPGMADRTVTISSAAKT
MLBr_01794|M.leprae_Br4923 LARRFGVRVISDEIHAPLVLSG---ARFTPYLSIPGAEN-AFALMSATKG
:* .: *::**:: *: . . * :.:: ** *
Mvan_5070|M.vanbaalenii_PYR-1 FNVTGWKIGWACGPSDLIAGVRAAKQYLSYVGGAPFQPAVAHALATEDAW
Mflv_1680|M.gilvum_PYR-GCK FNVTGWKIGWACGPSDLIAGVRAAKQYLSYVAGAPFQPAVAQALNNEDAW
MSMEG_5725|M.smegmatis_MC2_155 FNATGWKIGWACGAPELIAGVRAAKQYLTYVGGAPFQPAVAHALNHEDGW
Mb0881c|M.bovis_AF2122/97 FNCTGWKIGWACGPAELIAGVRAAKQYLSYVGGAPFQPAVALALDTEDAW
Rv0858c|M.tuberculosis_H37Rv FNCTGWKIGWACGPAELIAGVRAAKQYLSYVGGAPFQPAVALALDTEDAW
MAV_0979|M.avium_104 FNCTGWKIGWACGPAHLIAGVRAAKQYLSYVGGAPFQPAVALALETQDAW
MMAR_4678|M.marinum_M FNCTGWKIGWACGPQELIAGVRAAKQYMSYVGGAPLQPAVALALDSEDAW
MUL_0297|M.ulcerans_Agy99 FNCTGWKIGWACGLKELIAGVRAAKQYMSYVGGAPLQPAVALALDSEDAW
TH_2032|M.thermoresistible__bu FNCTGWKIGWACGPSDLIAGVRAAKQYLSYVGGAPFQPAVAHALDHEDAW
MAB_0847c|M.abscessus_ATCC_199 FNCTGWKIGWACGTPELVAGVRAAKQFLSYVGGGPFQPAVAVALDTEQSW
MLBr_01794|M.leprae_Br4923 WNLCGLKAAIAIAGREAAADLRRIPKEVSHGPSHLGIIAHTAAFRTGSSW
:* * * . * . . *.:* : ::: . * : *: ..*
Mvan_5070|M.vanbaalenii_PYR-1 VDALCASFQSRRDRLGSALSD--IGFEVHESFGTYFLCVDPRPLGFTDSA
Mflv_1680|M.gilvum_PYR-GCK VDALCASFQARRDRLSSALSD--IGFDVFDSFGTYFLTVDPRPLGYDDST
MSMEG_5725|M.smegmatis_MC2_155 VADLRDSFQSKRDRLGSALAE--IGFEVHDSRGTYFLCADPRPLGYDDST
Mb0881c|M.bovis_AF2122/97 VAALRNSLRARRDRLAAGLTE--IGFAVHDSYGTYFLCADPRPLGYDDST
Rv0858c|M.tuberculosis_H37Rv VAALRNSLRARRDRLAAGLTE--IGFAVHDSYGTYFLCADPRPLGYDDST
MAV_0979|M.avium_104 VADLRDRLRARRDRLAAGLID--IGFEVHDSAGTYFLCADPRPLGYDDSV
MMAR_4678|M.marinum_M VAALRTSLQARRDRLAAGLSE--IGFGVHDSHGTYFLCADPRPLGYDDST
MUL_0297|M.ulcerans_Agy99 VVALRTSLQARRDRLAAGLSE--IGFGVHDSHGTYFLCADPRPLGYDDST
TH_2032|M.thermoresistible__bu VAALRDSLQSRRDTLAAALTE--IGFEVHDSAGTYFLCADPRPLGYPDST
MAB_0847c|M.abscessus_ATCC_199 VKTLRESLQRRRDRLSAALTG--LGFEVHSSDGGYFVCADPRPLGFSDSA
MLBr_01794|M.leprae_Br4923 LDALLQGLEANRALLRDLVTKHLPGVRYQRPQGTYLTWLDCRHLGFDEQI
: * :. .* * : *. . * *: * * **: :.
Mvan_5070|M.vanbaalenii_PYR-1 AFCAELPERVGVAAIPMSAFCDPAAG----HAEEWKHLVRFAFCKREDTL
Mflv_1680|M.gilvum_PYR-GCK SFCAQLPEQVGVAAIPMSAFCDPDAA----HAGEWKHLVRFAFCKRDETL
MSMEG_5725|M.smegmatis_MC2_155 AFCAELPTRAGVAAIPMSAFCDPDAP----HADVWNHLVRFAFCKRDDTL
Mb0881c|M.bovis_AF2122/97 EFCAALPEKVGVAAIPMSAFCDPAAGQASQQADVWNHLVRFTFCKRDDTL
Rv0858c|M.tuberculosis_H37Rv EFCAALPEKVGVAAIPMSAFCDPAAGQASQQADVWNHLVRFTFCKRDDTL
MAV_0979|M.avium_104 AFCAALPEKVGVAAIPLSPFCDPAG--AHGPADVWNHLVRFTFCKRDDTL
MMAR_4678|M.marinum_M AFCAALPEKVGVAAIPMSAFCDPVAEHASTQADIWNHLVRFTFCKREDTL
MUL_0297|M.ulcerans_Agy99 AFCAALPERVGVAAIPMSAFCDPVAEHAPTQADIWNHLVRFTFCKREDTL
TH_2032|M.thermoresistible__bu EFCAGLPERVGVAAIPMSAFCDPAAP----HAEVWNHLVRFTFCKRPETL
MAB_0847c|M.abscessus_ATCC_199 ELCRRLPETVGVAAVPVSAFCDRYA-------DQWNHLVRFTFAKRDVVI
MLBr_01794|M.leprae_Br4923 NEGLAAISDLSGPARWFLDHSRVALTSGHAFGTGGVGHVRVNFATSQAIL
. .* . .. **. *.. :
Mvan_5070|M.vanbaalenii_PYR-1 DEAIRRLRALPRSR--
Mflv_1680|M.gilvum_PYR-GCK DEAIRRLQALRSTG--
MSMEG_5725|M.smegmatis_MC2_155 DEAIRRLQALRRC---
Mb0881c|M.bovis_AF2122/97 DEAIRRLSVLAERPAT
Rv0858c|M.tuberculosis_H37Rv DEAIRRLSVLAERPAT
MAV_0979|M.avium_104 DEAIRRLAALRGTG--
MMAR_4678|M.marinum_M DEAIKRLAALRDAA--
MUL_0297|M.ulcerans_Agy99 DEAIKRLAALRDAA--
TH_2032|M.thermoresistible__bu EEGIRRLGALRRG---
MAB_0847c|M.abscessus_ATCC_199 DEAITRLARLGA----
MLBr_01794|M.leprae_Br4923 IEALSRMGRALVNFR-
*.: *: