For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVSRLQPYATTVFAEMSALAARIGAVNLGQGFPDEDGPPAMLKAAQEAIAAGANQYPPGMGIPALRQAI AAQRRRRFGVEYDPDTEVLVTVGATEAIAAAVLGLVEPGSEVLLIEPFYDSYAPVIAMAGAKRVAVPLVP DGRGFALDADALRRAVTPKTRALIVNSPHNPTGAVLRRAELAAIAEIAVAADLLVITDEVYEHLVFTSPT GHQHLPLAGFDGMAQRTVTISSAAKMFNCTGWKIGWACGPQELIAGVRAAKQYMSYVGGAPLQPAVALAL DSEDAWVAALRTSLQARRDRLAAGLSEIGFGVHDSHGTYFLCADPRPLGYDDSTAFCAALPEKVGVAAIP MSAFCDPVAEHASTQADIWNHLVRFTFCKREDTLDEAIKRLAALRDAA
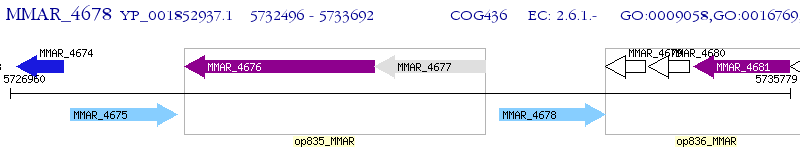
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4678 | - | - | 100% (398) | aminotransferase |
| M. marinum M | MMAR_5054 | aspB | 3e-32 | 29.97% (367) | aspartate aminotransferase AspB |
| M. marinum M | MMAR_4273 | - | 2e-22 | 27.76% (389) | aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0881c | - | 0.0 | 85.86% (396) | aminotransferase |
| M. gilvum PYR-GCK | Mflv_1680 | - | 1e-171 | 75.95% (395) | aminotransferase |
| M. tuberculosis H37Rv | Rv0858c | - | 0.0 | 85.86% (396) | aminotransferase |
| M. leprae Br4923 | MLBr_01488 | - | 5e-21 | 28.75% (320) | N-succinyldiaminopimelate aminotransferase |
| M. abscessus ATCC 19977 | MAB_0847c | - | 1e-153 | 68.45% (393) | aminotransferase |
| M. avium 104 | MAV_0979 | - | 0.0 | 83.80% (395) | aminotransferase |
| M. smegmatis MC2 155 | MSMEG_5725 | - | 1e-175 | 77.22% (395) | aminotransferase |
| M. thermoresistible (build 8) | TH_2032 | - | 0.0 | 81.77% (395) | PLP-dependent aminotransferases |
| M. ulcerans Agy99 | MUL_0297 | - | 0.0 | 98.24% (398) | aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_5070 | - | 1e-175 | 77.41% (394) | aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1680|M.gilvum_PYR-GCK -----MTVQRLRPYAVTIFAETSALAARVGAVNLGQGFPDEDGPPAMLKA
Mvan_5070|M.vanbaalenii_PYR-1 -----MTVRRLQPYAVTIFAEMSALAARVGAVNLGQGFPDEDGPPAMLKA
MSMEG_5725|M.smegmatis_MC2_155 -----MTVRRLQPFAVTIFAEMSALATRLGAVNLGQGFPDEDGPVEMLKV
MMAR_4678|M.marinum_M -----MTVSRLQPYATTVFAEMSALAARIGAVNLGQGFPDEDGPPAMLKA
MUL_0297|M.ulcerans_Agy99 -----MTVSRLQPYATTVFAEMSALAARIGAVNLGQGFPDEDGPPAMLKA
Mb0881c|M.bovis_AF2122/97 -----MTVSRLRPYATTVFAEMSALATRIGAVNLGQGFPDEDGPPKMLQA
Rv0858c|M.tuberculosis_H37Rv -----MTVSRLRPYATTVFAEMSALATRIGAVNLGQGFPDEDGPPKMLQA
MAV_0979|M.avium_104 -----MTVSRLRPYATTVFAEMSALAARVEAVNLGQGFPDEDGPAEMLKA
TH_2032|M.thermoresistible__bu -----MTVERLRPYATTIFAEMSALAARVGAVNLGQGFPDEDGPPAMLEI
MAB_0847c|M.abscessus_ATCC_199 --MNQRTVERLRPFGATIFAEMSALAVRHDAINLGQGFPDEDGPASMLDA
MLBr_01488|M.leprae_Br4923 MSPFGRVSATLPVFPWDTLADAKAVAESHPDGIVDLSVGTPVDPVAPLIQ
. * : :*: .*:* :. .. .* *
Mflv_1680|M.gilvum_PYR-GCK AENAIAEGVNQYPPGLGFAPLRQAIADQRRRRYGTVYDPDTEVLVTVGAT
Mvan_5070|M.vanbaalenii_PYR-1 AENAIAEGVNQYPPGLGIAPLRQAIADQRRRRYGSEYDPDTEVLVTVGAT
MSMEG_5725|M.smegmatis_MC2_155 AQNAIADGINQYPPGLGVPELRQAIAAQRKRVHGVQYDPDTEVLVTVGAT
MMAR_4678|M.marinum_M AQEAIAAGANQYPPGMGIPALRQAIAAQRRRRFGVEYDPDTEVLVTVGAT
MUL_0297|M.ulcerans_Agy99 AQEAIAAGANQYPPGMGIPALRQAIAAQRRRRFGVEYDPDTEVLVTVGAT
Mb0881c|M.bovis_AF2122/97 AQDAIAGGVNQYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGAT
Rv0858c|M.tuberculosis_H37Rv AQDAIAGGVNQYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGAT
MAV_0979|M.avium_104 AQEAIADGVNQYPPGIGIAPLRHAIAAQRHRRYGIEYDPDTEVLVTVGAT
TH_2032|M.thermoresistible__bu ARDAIAEGMNQYPPGIGIAPLRQAIAAQRERHYGVEYDPDTEVLVTVGAT
MAB_0847c|M.abscessus_ATCC_199 AQQAIRSGLNQYPPGLGIPELRRAITADRLARYGEELDPDTQVLVTVGAT
MLBr_01488|M.leprae_Br4923 EALAAASSVAGYPTTAGTARLREAAVAALDRRYGITGLTEAAVLPVIGTK
* . **. * . **.* . .* .:: ** .:*:.
Mflv_1680|M.gilvum_PYR-GCK EAIASSVLGL-VEPGSEVLLIEPFYDSYSPVIAMAGCHRRAVPLRQNGRG
Mvan_5070|M.vanbaalenii_PYR-1 EAIAAAILGL-VEPDSEVLLIEPFYDSYSPVIAMAGCHRRAVPMVRDGRG
MSMEG_5725|M.smegmatis_MC2_155 EAIAASVIGL-VEPGSEVLVIEPFYDSYSPVIAMAGCERRAVPLVPSGRG
MMAR_4678|M.marinum_M EAIAAAVLGL-VEPGSEVLLIEPFYDSYAPVIAMAGAKRVAVPLVPDGRG
MUL_0297|M.ulcerans_Agy99 EAIAAAVRGL-VEPGSEVLLIEPFYDSYAPVIAMAGAKRVAVPLVPDGRG
Mb0881c|M.bovis_AF2122/97 EAIAAAVLGL-VEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRG
Rv0858c|M.tuberculosis_H37Rv EAIAAAVLGL-VEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRG
MAV_0979|M.avium_104 EAIASAVIGL-VEPGSEVLLIEPFYDSYSPVIAMASAQRVAVPLVPHGRG
TH_2032|M.thermoresistible__bu EAIAGAVLGL-VEPGSEVLLIEPFYDSYSPVIAMAGAHRVAVPLVTDGPG
MAB_0847c|M.abscessus_ATCC_199 EAIAGALLGL-VEPGSEVILIEPYYDSYAAVVAMAGAVRVPVPLVPDGAG
MLBr_01488|M.leprae_Br4923 ELIAWLPTLLGLRAGDVVVVPELAYPTYNVGARLAGAR------------
* ** * :.... *:: * * :* :*..
Mflv_1680|M.gilvum_PYR-GCK FAIDIEALRAAVGPRTRALIVNTPHNPTGMIASDEELRGLAELAVAHDLL
Mvan_5070|M.vanbaalenii_PYR-1 FAIDVEGLRRAVTPKTRALILNSPHNPTGSVASDDELRAVAELAISADLL
MSMEG_5725|M.smegmatis_MC2_155 FAIDVDALRAAVTPQTKALILNSPHNPTGAVASDAELRAVAALAVEHDLL
MMAR_4678|M.marinum_M FALDADALRRAVTPKTRALIVNSPHNPTGAVLRRAELAAIAEIAVAADLL
MUL_0297|M.ulcerans_Agy99 FALDADALRRAVTPKTRALIVNSPHNPTGAVLSRAELAAIAEIAVAADLL
Mb0881c|M.bovis_AF2122/97 FALDADALRRAVTPRTRALIINSPHNPTGAVLSATELAAIAEIAVAANLV
Rv0858c|M.tuberculosis_H37Rv FALDADALRRAVTPRTRALIINSPHNPTGAVLSATELAAIAEIAVAANLV
MAV_0979|M.avium_104 FALDADALRRAVTPRTRALIVNSPHNPTGTVLGATELAAIAEVAVESDLL
TH_2032|M.thermoresistible__bu FAVDLDALRRAVTPRTRAMIVNSPHNPTGMVLTEAELAGIAELAVSADLL
MAB_0847c|M.abscessus_ATCC_199 FALDTDALAAAITPRTAALVINSPHNPTGKVFTDTELARIAELAVEHDLL
MLBr_01488|M.leprae_Br4923 -VLRADSLTQLGLQSPALFYLNSPSNPTGQVLAVNHLRKMVGWARNRGVV
.: :.* . : :*:* **** : .* :. * .::
Mflv_1680|M.gilvum_PYR-GCK VITDEVYEHLVFDG---RQHRPLAGYPGMAERTVTISSAGKMFNVTGWKI
Mvan_5070|M.vanbaalenii_PYR-1 VITDEVYEHLVFDG---RRHRPLADYPGMAGRTITISSAGKMFNVTGWKI
MSMEG_5725|M.smegmatis_MC2_155 VITDEVYEALVFDG---HRHLTLASYPGMRDRTVTISSAAKMFNATGWKI
MMAR_4678|M.marinum_M VITDEVYEHLVFTSPTGHQHLPLAGFDGMAQRTVTISSAAKMFNCTGWKI
MUL_0297|M.ulcerans_Agy99 VITDEVYEHLVFTSPTGHQHLPLAGFDGMAQRTVTISSAAKMFNCTGWKI
Mb0881c|M.bovis_AF2122/97 VITDEVYEHLVFDH---ARHLPLAGFDGMAERTITISSAAKMFNCTGWKI
Rv0858c|M.tuberculosis_H37Rv VITDEVYEHLVFDH---ARHLPLAGFDGMAERTITISSAAKMFNCTGWKI
MAV_0979|M.avium_104 VITDEVYEHLVFDD---HRHLPLAGFDGMAERTITISSAAKMFNCTGWKI
TH_2032|M.thermoresistible__bu VITDEVYEHLVYPSPAGRRHIPLASYPGMAERTVTISSAAKMFNCTGWKI
MAB_0847c|M.abscessus_ATCC_199 VVSDEVYERLLFDG---RTQTPMASLPGMADRTVTISSAAKTFNCTGWKI
MLBr_01488|M.leprae_Br4923 VVSDECYLGLGWDAEPFSVLHPLV-CDGNHTGLLAVHSLSKSSSLAGYRA
*::** * * : .:. * ::: * .* . :*::
Mflv_1680|M.gilvum_PYR-GCK GWACGPSDLIAGVRAAKQYLSYVAGAPFQPAVAQALNNEDAWVDALCASF
Mvan_5070|M.vanbaalenii_PYR-1 GWACGPSDLIAGVRAAKQYLSYVGGAPFQPAVAHALATEDAWVDALCASF
MSMEG_5725|M.smegmatis_MC2_155 GWACGAPELIAGVRAAKQYLTYVGGAPFQPAVAHALNHEDGWVADLRDSF
MMAR_4678|M.marinum_M GWACGPQELIAGVRAAKQYMSYVGGAPLQPAVALALDSEDAWVAALRTSL
MUL_0297|M.ulcerans_Agy99 GWACGLKELIAGVRAAKQYMSYVGGAPLQPAVALALDSEDAWVVALRTSL
Mb0881c|M.bovis_AF2122/97 GWACGPAELIAGVRAAKQYLSYVGGAPFQPAVALALDTEDAWVAALRNSL
Rv0858c|M.tuberculosis_H37Rv GWACGPAELIAGVRAAKQYLSYVGGAPFQPAVALALDTEDAWVAALRNSL
MAV_0979|M.avium_104 GWACGPAHLIAGVRAAKQYLSYVGGAPFQPAVALALETQDAWVADLRDRL
TH_2032|M.thermoresistible__bu GWACGPSDLIAGVRAAKQYLSYVGGAPFQPAVAHALDHEDAWVAALRDSL
MAB_0847c|M.abscessus_ATCC_199 GWACGTPELVAGVRAAKQFLSYVGGGPFQPAVAVALDTEQSWVKTLRESL
MLBr_01488|M.leprae_Br4923 GFVAGDPDLVAELLAVRKHAGMMVPTPVQAAMVAALG-DDAHEREQRDRY
*:..* .*:* : *.::. : *.*.*:. ** ::.
Mflv_1680|M.gilvum_PYR-GCK QARRDRLSSALSDIGFDVFDSFGTYFLTVDPRPLGYDDSTSFCAQLPEQV
Mvan_5070|M.vanbaalenii_PYR-1 QSRRDRLGSALSDIGFEVHESFGTYFLCVDPRPLGFTDSAAFCAELPERV
MSMEG_5725|M.smegmatis_MC2_155 QSKRDRLGSALAEIGFEVHDSRGTYFLCADPRPLGYDDSTAFCAELPTRA
MMAR_4678|M.marinum_M QARRDRLAAGLSEIGFGVHDSHGTYFLCADPRPLGYDDSTAFCAALPEKV
MUL_0297|M.ulcerans_Agy99 QARRDRLAAGLSEIGFGVHDSHGTYFLCADPRPLGYDDSTAFCAALPERV
Mb0881c|M.bovis_AF2122/97 RARRDRLAAGLTEIGFAVHDSYGTYFLCADPRPLGYDDSTEFCAALPEKV
Rv0858c|M.tuberculosis_H37Rv RARRDRLAAGLTEIGFAVHDSYGTYFLCADPRPLGYDDSTEFCAALPEKV
MAV_0979|M.avium_104 RARRDRLAAGLIDIGFEVHDSAGTYFLCADPRPLGYDDSVAFCAALPEKV
TH_2032|M.thermoresistible__bu QSRRDTLAAALTEIGFEVHDSAGTYFLCADPRPLGYPDSTEFCAGLPERV
MAB_0847c|M.abscessus_ATCC_199 QRRRDRLSAALTGLGFEVHSSDGGYFVCADPRPLGFSDSAELCRRLPETV
MLBr_01488|M.leprae_Br4923 ARRRDVLLPAVVSAGFTVDHSEAGLYLWAS-RGEPCRDSVEWFAQR----
:** * ..: ** * * . :: .. * **.
Mflv_1680|M.gilvum_PYR-GCK GVAAIPMSAFCDPDAA----HAGEWKHLVRFAFCKRDETLDEAIRRLQAL
Mvan_5070|M.vanbaalenii_PYR-1 GVAAIPMSAFCDPAAG----HAEEWKHLVRFAFCKREDTLDEAIRRLRAL
MSMEG_5725|M.smegmatis_MC2_155 GVAAIPMSAFCDPDAP----HADVWNHLVRFAFCKRDDTLDEAIRRLQAL
MMAR_4678|M.marinum_M GVAAIPMSAFCDPVAEHASTQADIWNHLVRFTFCKREDTLDEAIKRLAAL
MUL_0297|M.ulcerans_Agy99 GVAAIPMSAFCDPVAEHAPTQADIWNHLVRFTFCKREDTLDEAIKRLAAL
Mb0881c|M.bovis_AF2122/97 GVAAIPMSAFCDPAAGQASQQADVWNHLVRFTFCKRDDTLDEAIRRLSVL
Rv0858c|M.tuberculosis_H37Rv GVAAIPMSAFCDPAAGQASQQADVWNHLVRFTFCKRDDTLDEAIRRLSVL
MAV_0979|M.avium_104 GVAAIPLSPFCDPAG--AHGPADVWNHLVRFTFCKRDDTLDEAIRRLAAL
TH_2032|M.thermoresistible__bu GVAAIPMSAFCDPAAP----HAEVWNHLVRFTFCKRPETLEEGIRRLGAL
MAB_0847c|M.abscessus_ATCC_199 GVAAVPVSAFCDRYA-------DQWNHLVRFTFAKRDVVIDEAITRLARL
MLBr_01488|M.leprae_Br4923 -GILVALGEFYGPGG----------CQHVRVALTASDERIAAVVARLS--
:.:. * . . : **.:: : : **
Mflv_1680|M.gilvum_PYR-GCK RSTG--
Mvan_5070|M.vanbaalenii_PYR-1 PRSR--
MSMEG_5725|M.smegmatis_MC2_155 RRC---
MMAR_4678|M.marinum_M RDAA--
MUL_0297|M.ulcerans_Agy99 RDAA--
Mb0881c|M.bovis_AF2122/97 AERPAT
Rv0858c|M.tuberculosis_H37Rv AERPAT
MAV_0979|M.avium_104 RGTG--
TH_2032|M.thermoresistible__bu RRG---
MAB_0847c|M.abscessus_ATCC_199 GA----
MLBr_01488|M.leprae_Br4923 ------