For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHSSAHHRQQRTFAQSSKLQDVLYEIRGPVHQHAARLEAEGHRILKLNIGNPAPFGFEAPDVIMRDMIQA LPYAQGYSDSQGILPARRAVVTRYELVDGFPRFDVDDVYLGNGVSELITMTLQALLDNGDEVLIPSPDYP LWTASTSLAGGTPVHYLCDETQGWQPDIADLESKITERTKALVVINPNNPTGAVYSGEILSQMVDLARKH ELLLLADEIYDKILYDAAKHINVASLAPDMLCLTFNGLSKAYRVAGYRAGWLAITGPKDHAGSFIEGINL LANMRLCPNVPAQHAIQVALGGHQSIEDLVLPGGRLLEQRDVAWSKLNEIPGVSCVKPAGALYAFPRLDP EVYDITDDEQLVLDLLLQEKILVTQGTGFNWPAPDHLRIVTLPWARDLAAAIERLGNFLVGYRQ
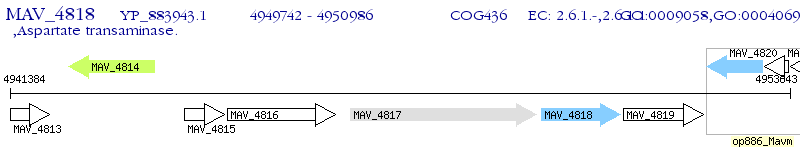
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4818 | - | - | 100% (414) | aminotransferase AlaT |
| M. avium 104 | MAV_0596 | - | 3e-20 | 25.65% (382) | aspartate aminotransferase |
| M. avium 104 | MAV_0979 | - | 3e-18 | 26.29% (232) | aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0344c | aspC | 0.0 | 93.43% (411) | aminotransferase AlaT |
| M. gilvum PYR-GCK | Mflv_0279 | - | 0.0 | 86.86% (411) | aminotransferase AlaT |
| M. tuberculosis H37Rv | Rv0337c | aspC | 0.0 | 93.43% (411) | aminotransferase AlaT |
| M. leprae Br4923 | MLBr_02502 | - | 0.0 | 91.55% (414) | aminotransferase AlaT |
| M. abscessus ATCC 19977 | MAB_4294 | - | 0.0 | 88.45% (407) | aminotransferase AlaT |
| M. marinum M | MMAR_0610 | aspC | 0.0 | 93.22% (413) | aspartate aminotransferase AspC |
| M. smegmatis MC2 155 | MSMEG_0688 | - | 0.0 | 88.59% (412) | aminotransferase AlaT |
| M. thermoresistible (build 8) | TH_0764 | aspC | 0.0 | 87.78% (409) | PROBABLE ASPARTATE AMINOTRANSFERASE ASPC (TRANSAMINASE A) |
| M. ulcerans Agy99 | MUL_0573 | aspC | 0.0 | 93.22% (413) | aminotransferase AlaT |
| M. vanbaalenii PYR-1 | Mvan_0618 | - | 0.0 | 88.32% (411) | aminotransferase AlaT |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0610|M.marinum_M --------MDSDETMVDVTTHQLPWHAS-SHQRQ-RSFAQSSKLQDVLYE
MUL_0573|M.ulcerans_Agy99 --------MDSDETMVDVTTHQLPWHAS-SHQRQ-RSFAQSSKLQDVLYE
Mb0344c|M.bovis_AF2122/97 --------MDNDGTIVDVTTHQLPWHTA-SHQRQ-RAFAQSAKLQDVLYE
Rv0337c|M.tuberculosis_H37Rv --------MDNDGTIVDVTTHQLPWHTA-SHQRQ-RAFAQSAKLQDVLYE
MLBr_02502|M.leprae_Br4923 MTLYQQISVDGSGTMSDVTTHQLPMHTP-GYHRQ-RAFAQSSKLQDVLYE
MAV_4818|M.avium_104 ------------------------MHSS-AHHRQQRTFAQSSKLQDVLYE
MAB_4294|M.abscessus_ATCC_1997 --------MSTDNLPIDVPP-RVPGLSP-RQHQR--VFAQSTKLQDVLYE
Mflv_0279|M.gilvum_PYR-GCK --------MSTHQAH----WQAGTGQH-----ARQREFAQSSKLQDVLYE
Mvan_0618|M.vanbaalenii_PYR-1 -----MSLVSTHQAH----WHTGSGQH-----ARQREFTQSSKLQDVLYE
MSMEG_0688|M.smegmatis_MC2_155 -----MERVTTHQLP----WQSTGEHR------KPRTFTQSTKLQDVLYE
TH_0764|M.thermoresistible__bu --------VTSHQLPAEKTWQGYSAHGPHGHPSRPRTFAQSSKLQDVLYE
: *:**:********
MMAR_0610|M.marinum_M IRGPVHQQAARLETEGHRILKLNIGNPAPFGFEAPDVIMRDMIQALPYAQ
MUL_0573|M.ulcerans_Agy99 IRGPVHQQAARLETEGHRILKLNIGNPAPFGFEAPDVIMRDMIQALPYAQ
Mb0344c|M.bovis_AF2122/97 IRGPVHQHAARLEAEGHRILKLNIGNPAPFGFEAPDVIMRDIIQALPYAQ
Rv0337c|M.tuberculosis_H37Rv IRGPVHQHAARLEAEGHRILKLNIGNPAPFGFEAPDVIMRDIIQALPYAQ
MLBr_02502|M.leprae_Br4923 IRGPVSQHATRLETEGHRILKLNIGNPALFGFDAPDVIMRDMIQALPYAQ
MAV_4818|M.avium_104 IRGPVHQHAARLEAEGHRILKLNIGNPAPFGFEAPDVIMRDMIQALPYAQ
MAB_4294|M.abscessus_ATCC_1997 IRGPVHAHAARLEAEGHRILKLNIGNPAPFGFEAPDVIMRDIIQALPYAQ
Mflv_0279|M.gilvum_PYR-GCK IRGPVHEHASRLEAEGHRILKLNIGNPAPFGFEAPDVIMRDIIAALPYAQ
Mvan_0618|M.vanbaalenii_PYR-1 IRGPIHEHASRLEAEGHRIFKLNIGNPAPFGFEAPDVIMRDIIAALPYAQ
MSMEG_0688|M.smegmatis_MC2_155 IRGPVHEHASRLEAEGHRILKLNIGNPAPFGFEAPDVIMRDIIQALPTAQ
TH_0764|M.thermoresistible__bu IRGPVHDHAARLEMEGHRILKLNIGNPAPFGFEAPDVIMRDIIQALPYAQ
****: :*:*** *****:******** ***:********:* *** **
MMAR_0610|M.marinum_M GYSDSQGILPARRAVVTRYELVDGFPRFDVDDVYLGNGVSELITMTLQAL
MUL_0573|M.ulcerans_Agy99 GYSDSQGILPARRAVVTRYELVDGFPRFDVDDVYLGNGVSELITMTLQAL
Mb0344c|M.bovis_AF2122/97 GYSDSQGILSARRAVVTRYELVPGFPRFDVDDVYLGNGVSELITMTLQAL
Rv0337c|M.tuberculosis_H37Rv GYSDSQGILSARRAVVTRYELVPGFPRFDVDDVYLGNGVSELITMTLQAL
MLBr_02502|M.leprae_Br4923 GYSDSQGILPARRAVVTRYELVDGFPRFDVDDVYLGNGVSELITMTLQAL
MAV_4818|M.avium_104 GYSDSQGILPARRAVVTRYELVDGFPRFDVDDVYLGNGVSELITMTLQAL
MAB_4294|M.abscessus_ATCC_1997 GYSDSKGILPARRAVVTRYELVDGFPYLDVDDVFLGNGVSELITMTTQAL
Mflv_0279|M.gilvum_PYR-GCK GYSDSKGIVSARRAVFTRYELVEGFPRFDIDDVFLGNGVSELIQMTLQAL
Mvan_0618|M.vanbaalenii_PYR-1 GYSDSKGIVSARRAVFTRYELVEGFPKFDIDDVFLGNGVSELITMTLQAL
MSMEG_0688|M.smegmatis_MC2_155 GYSDSKGILSARRAVFTRYELVEGFPKFDVDDVYLGNGVSELITMTLQAL
TH_0764|M.thermoresistible__bu GYSDSKGILPARRAVVTRYELVEGFPPFDVDDVYLGNGASELIQMTLQAL
*****:**:.*****.****** *** :*:***:****.**** ** ***
MMAR_0610|M.marinum_M LDNGDQVLIPAPDYPLWTASTSLAGGTPVHYLCDETQDWQPDIADLESKI
MUL_0573|M.ulcerans_Agy99 LDNGDQVLIPAPDYPLWTASTSLAGGTPVHYLCDETQDWQPDIADLESKI
Mb0344c|M.bovis_AF2122/97 LDNGDQVLIPSPDYPLWTASTSLAGGTPVHYLCDETQGWQPDIADLESKI
Rv0337c|M.tuberculosis_H37Rv LDNGDQVLIPSPDYPLWTASTSLAGGTPVHYLCDETQGWQPDIADLESKI
MLBr_02502|M.leprae_Br4923 LDNGDQVLIPSPDYPLWTASTSLAGGTPVHYLCDETQGWQPDIADVESKI
MAV_4818|M.avium_104 LDNGDEVLIPSPDYPLWTASTSLAGGTPVHYLCDETQGWQPDIADLESKI
MAB_4294|M.abscessus_ATCC_1997 LDNGDQVLIPAPDYPLWTAATSLAGGTAVHYLCDETNGWMPDIEDLESKI
Mflv_0279|M.gilvum_PYR-GCK LDNGDQVLIPAPDYPLWTACTSLAGGTPVHYLCDETQGWNPDVADIESKI
Mvan_0618|M.vanbaalenii_PYR-1 LDNGDQVLIPAPDYPLWTASTALAGGTPVHYLCDETQGWNPDIADLESKI
MSMEG_0688|M.smegmatis_MC2_155 LDNGDQVLIPAPDYPLWTASTSLAGGTPVHYLCDETQGWNPDVADIESKI
TH_0764|M.thermoresistible__bu LDNGDQVLIPAPDYPLWTACTSLAGGTPVHYLCDETQGWNPDIEDLESKI
*****:****:********.*:*****.********:.* **: *:****
MMAR_0610|M.marinum_M TERTKALVIINPNNPTGAVYSHEILTQMVELARKHELLLLADEIYDKILY
MUL_0573|M.ulcerans_Agy99 TERTKALVIINPNNPTGAVYSHEILTQMVELARKHELLLLADEIYDKILY
Mb0344c|M.bovis_AF2122/97 TERTKALVVINPNNPTGAVYSCEILTQMVDLARKHQLLLLADEIYDKILY
Rv0337c|M.tuberculosis_H37Rv TERTKALVVINPNNPTGAVYSCEILTQMVDLARKHQLLLLADEIYDKILY
MLBr_02502|M.leprae_Br4923 TERTKALVVINPNNPTGAVYSNEILNQIVDLARKHQLLLLADEIYDKILY
MAV_4818|M.avium_104 TERTKALVVINPNNPTGAVYSGEILSQMVDLARKHELLLLADEIYDKILY
MAB_4294|M.abscessus_ATCC_1997 TERTKALVIINPNNPTGAVYSREILTKMVELARKHQLLLLADEIYDKILY
Mflv_0279|M.gilvum_PYR-GCK TDRTKAIVVINPNNPTGAVYSREVLTQIADLARKHQLLLLADEIYDKILY
Mvan_0618|M.vanbaalenii_PYR-1 TERTKALVVINPNNPTGAVYSRETLEQMVELARKHQLLLLADEIYDKILY
MSMEG_0688|M.smegmatis_MC2_155 TERTKAIVVINPNNPTGAVYSRETLEQIVDLARKHQLLLLADEIYDKILY
TH_0764|M.thermoresistible__bu TERTKAIVVINPNNPTGAVYTRETLEQIAELARRHQLLLLADEIYDKILY
*:****:*:***********: * * ::.:***:*:**************
MMAR_0610|M.marinum_M DDAKHISVATLAPDMLCLTFNGLSKAYRVAGYRAGWLAITGPKEHASSFI
MUL_0573|M.ulcerans_Agy99 DDAKHISVATLAPDMLCLTFNGLSKAYRVAGYRAGWLAITGPKEHASSFI
Mb0344c|M.bovis_AF2122/97 DDAKHISLASIAPDMLCLTFNGLSKAYRVAGYRAGWLAITGPKEHASSFI
Rv0337c|M.tuberculosis_H37Rv DDAKHISLASIAPDMLCLTFNGLSKAYRVAGYRAGWLAITGPKEHASSFI
MLBr_02502|M.leprae_Br4923 DDTKHISVASIAPDLLCLTFNGLSKAYRVAGYRSGWLAITGPKDHASSFI
MAV_4818|M.avium_104 DAAKHINVASLAPDMLCLTFNGLSKAYRVAGYRAGWLAITGPKDHAGSFI
MAB_4294|M.abscessus_ATCC_1997 DDAEHISVASLAPDLLCFTFNGLSKAYRVAGYRSAWLAITGPKDHAASLL
Mflv_0279|M.gilvum_PYR-GCK DDAEHISLASVAPDLLTLTFNGLSKAYRVAGYRSGWLVITGPKEHAASFI
Mvan_0618|M.vanbaalenii_PYR-1 DDAKHISLATLAPDLLTLTFNGLSKAYRVAGYRSGWLVITGPKEHAASFI
MSMEG_0688|M.smegmatis_MC2_155 DDAKHISLATLAPDLLCLTFNGLSKAYRVAGYRSGWLVITGPKEHATSFI
TH_0764|M.thermoresistible__bu DDAKHISMAAVAPDVLTLTFNGLSKAYRVAGYRSGWLCITGPKEHAASFI
* ::**.:*::***:* :***************:.** *****:** *::
MMAR_0610|M.marinum_M EGISLLANMRLCPNVPAQHAIQVALGGHQSIDDLVLPGGRLLEQRDVAWS
MUL_0573|M.ulcerans_Agy99 EGISLLANMRLCPNVPAQHAIQVALGGHQSIDDLVLPGGRLLEQRDVAWS
Mb0344c|M.bovis_AF2122/97 EGIGLLANMRLCPNVPAQHAIQVALGGHQSIEDLVLPGGRLLEQRDIAWT
Rv0337c|M.tuberculosis_H37Rv EGIGLLANMRLCPNVPAQHAIQVALGGHQSIEDLVLPGGRLLEQRDIAWT
MLBr_02502|M.leprae_Br4923 GGINLLANMRLCPNVPAQHAIQVALGGHQSIEDLVLPGGRLLEQRDVAWT
MAV_4818|M.avium_104 EGINLLANMRLCPNVPAQHAIQVALGGHQSIEDLVLPGGRLLEQRDVAWS
MAB_4294|M.abscessus_ATCC_1997 EGVNLLANMRLCPNVPAQHAIQVALGGHQSIEDLVLPGGRLLEQRDVAWT
Mflv_0279|M.gilvum_PYR-GCK EGISLLANMRLCPNVPAQHAIQVALGGHQSIDDLVLPGGRLLEQRDIAWE
Mvan_0618|M.vanbaalenii_PYR-1 EGINLLANMRLCPNVPAQHAIQVALGGHQSIDDLVLPGGRLLEQRDVAWE
MSMEG_0688|M.smegmatis_MC2_155 EGISLLANMRLCPNVPAQHAIQVALGGHQSIDALVLPGGRLLEQRDVAWE
TH_0764|M.thermoresistible__bu EGISLLANMRLCPNVPAQHAIQVALGGHQSIEELVLPGGRLREQRDVAWE
*:.***************************: ******** ****:**
MMAR_0610|M.marinum_M KLNEIPGVSCVKPKGALYAFPRLDPEVYDIADDEQLVLDLLLSEKILVTQ
MUL_0573|M.ulcerans_Agy99 KLNEIPGVSCVKPKGALYAFPRLDPEVYDIADDEQLVLDLLLSEKILVTQ
Mb0344c|M.bovis_AF2122/97 KLNEIPGVSCVKPAGALYAFPRLDPEVYDIDDDEQLVLDLLLSEKILVTQ
Rv0337c|M.tuberculosis_H37Rv KLNEIPGVSCVKPAGALYAFPRLDPEVYDIDDDEQLVLDLLLSEKILVTQ
MLBr_02502|M.leprae_Br4923 KLNEIPGVSCVKPTGALYAFPRLDPEVYGIVDDEQLVLDLLLQEKILVTQ
MAV_4818|M.avium_104 KLNEIPGVSCVKPAGALYAFPRLDPEVYDITDDEQLVLDLLLQEKILVTQ
MAB_4294|M.abscessus_ATCC_1997 KLNEIPGVSCVKPQGALYAFPRLDPEVHHIHDDDQLVLDLLLNEKILLTQ
Mflv_0279|M.gilvum_PYR-GCK TLNEIPGVSCVKPQGALYAFPRLDPEVYDIADDEQLVLDLLLQEKILVVQ
Mvan_0618|M.vanbaalenii_PYR-1 KLNEIPGVSCVKPKGALYAFPRLDPEVYDVEDDEQLVLDLLLQEKILLTQ
MSMEG_0688|M.smegmatis_MC2_155 KLNEIPGVSCVKPRGALYAFPRLDPEVYDIHDDEQLVLDLLLQEKILLTQ
TH_0764|M.thermoresistible__bu KLNEIPGVSCVKPQGALYVFPRLDPEVYEIHDDEQLVLDLLLQEKILLTQ
.************ ****.********: : **:********.****:.*
MMAR_0610|M.marinum_M GTGFNWPEPDHLRLVTLPWSRDLAAAIERLGNFLVSYRQ-
MUL_0573|M.ulcerans_Agy99 GTGFNWPEPDHLRLVTLPWSRDLAAAIERLGNFLVSYRQ-
Mb0344c|M.bovis_AF2122/97 GTGFNWPAPDHLRLVTLPWSRDLAAAIERLGNFLVSYRQ-
Rv0337c|M.tuberculosis_H37Rv GTGFNWPAPDHLRLVTLPWSRDLAAAIERLGNFLVSYRQ-
MLBr_02502|M.leprae_Br4923 GSGFNWPAPDHLRLVTLPWSRDLAAAIDRLGNFLVSYRQ-
MAV_4818|M.avium_104 GTGFNWPAPDHLRIVTLPWARDLAAAIERLGNFLVGYRQ-
MAB_4294|M.abscessus_ATCC_1997 GTGFNWPEPDHLRIVTLPWARDLAVAIERLGNFLASYRQ-
Mflv_0279|M.gilvum_PYR-GCK GTGFNWPTPDHLRIVTLPWARDLSAAIERLGNFLVSYRQ-
Mvan_0618|M.vanbaalenii_PYR-1 GTGFNWPTPDHLRIVTLPWSRDLAAAIERLGNFLVSYRQQ
MSMEG_0688|M.smegmatis_MC2_155 GTGFNWPTPDHLRIVTLPWARDLASAIERLGNFLASYRQ-
TH_0764|M.thermoresistible__bu GTGFNWPTPDHLRIVTLPWARDLANAIERLGNFLVSYRQ-
*:***** *****:*****:***: **:******..***