For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSVHVTDAKPVSEAEQLLEGLNPQQRQAVVHEGSPLLIVAGAGSGKTAVLTRRVAYLIAARGVGVGQVLA ITFTNKAAAEMRERVVRLVGNRARAMWVSTFHSTCVRILRNQASLIEGLNSNFSIYDADDSRRLLQMIGR DMGLDIKRYSPRLLANAISNLKNELIDPAAAVANLTDDSDELSRTVASVYGEYQRRLRAANALDFDDLIG ETVAVLRAFPQIAQHYRRRFRHVLVDEYQDTNHAQYVLVRELVGTDTEDGVPPGELCVVGDADQSIYAFR GATIRNIEDFERDYPDATTILLEQNYRSTQNILSAANSVIARNSGRRDKRLWTDAGAGELIVGYVADNEH DEARFVAEEIDALAQKGEITYNDVAVFYRTNNSSRSFEEVFIRAGIPYKVVGGVRFYERKEIRDIIAYLR VLDNPGDAVSMRRILNTPRRGIGDRAEACVAVYAENTGASFADALVAAAEGKVPMLNSRAEKAIAGFVEL LDDLRGRLDDDLGDLVESVLERTGYRRELESSTDPQELARLDNLNELVSVAHEFSTDRANAAALDESLDA ADDEDVPDTGVLAEFLERVSLVSDSDEIPEDGAGMVTLMTLHTAKGLEFPVVFVTGWEDGMFPHMRSLDD PVELSEERRLAYVGITRARQRLYLSRAIVRSSWGQPMLNPESRFLREIPQELIEWRRTAPAPSFSAPVSG AGRFGTPRAAPTRAAMSKRPLLVLAPGDRVTHDKYGLGRVEEVSGAGESAMSLIDFGSSGRVKLMHNHAP ISKL
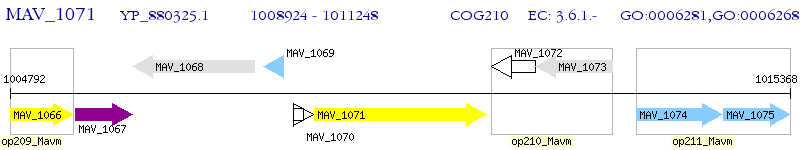
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1071 | pcrA | - | 100% (774) | ATP-dependent DNA helicase PcrA |
| M. avium 104 | MAV_4143 | - | 4e-57 | 35.29% (527) | ATP-dependent DNA helicase |
| M. avium 104 | MAV_4147 | - | 1e-46 | 26.00% (873) | superfamily protein I DNA and RNA helicases |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0974 | uvrD1 | 0.0 | 88.92% (776) | ATP dependent DNA helicase UVRD1 |
| M. gilvum PYR-GCK | Mflv_1861 | - | 0.0 | 83.61% (781) | ATP-dependent DNA helicase PcrA |
| M. tuberculosis H37Rv | Rv0949 | uvrD1 | 0.0 | 88.92% (776) | ATP-dependent DNA helicase II UVRD1 |
| M. leprae Br4923 | MLBr_00153 | uvrD | 0.0 | 82.65% (778) | putative ATP-dependent DNA helicase |
| M. abscessus ATCC 19977 | MAB_1054 | - | 0.0 | 74.72% (791) | putative ATP-dependent DNA helicase PcrA |
| M. marinum M | MMAR_4553 | uvrD1 | 0.0 | 88.46% (780) | ATP-dependent DNA helicase II UvrD1 |
| M. smegmatis MC2 155 | MSMEG_5534 | pcrA | 0.0 | 81.89% (773) | ATP-dependent DNA helicase PcrA |
| M. thermoresistible (build 8) | TH_1135 | uvrD2 | 2e-55 | 36.78% (435) | PROBABLE ATP-DEPENDENT DNA HELICASE II UVRD2 |
| M. ulcerans Agy99 | MUL_4723 | uvrD1 | 0.0 | 87.69% (780) | ATP-dependent DNA helicase II UvrD1 |
| M. vanbaalenii PYR-1 | Mvan_4873 | - | 0.0 | 84.65% (762) | ATP-dependent DNA helicase PcrA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1861|M.gilvum_PYR-GCK -MTAHVTLPE-ATPMAG----DDLLEGLNPQQRQAVLHEGTPLLIVAGAG
Mvan_4873|M.vanbaalenii_PYR-1 -MTSLSVS---AGPTGT----DELLDGLNPQQRQAVLHQGTPLLIVAGAG
MSMEG_5534|M.smegmatis_MC2_155 -MTSPSAT---PFAAET----DQLLDGLNPQQRQAVLHEGSPLLIVAGAG
MMAR_4553|M.marinum_M -MTVQATDA------------DQLLEGLNPQQRQAVVHEGSPLLIVAGAG
MUL_4723|M.ulcerans_Agy99 -MTVQATDA------------DQLVEGLNPQQRQAVVHEGSPLLIVAGAG
Mb0974|M.bovis_AF2122/97 -MSVHATDAKPPGPSPA----DQLLDGLNPQQRQAVVHEGSPLLIVAGAG
Rv0949|M.tuberculosis_H37Rv -MSVHATDAKPPGPSPA----DQLLDGLNPQQRQAVVHEGSPLLIVAGAG
MAV_1071|M.avium_104 -MSVHVTDAKP--VSEA----EQLLEGLNPQQRQAVVHEGSPLLIVAGAG
MLBr_00153|M.leprae_Br4923 -MSVHTTDAKP--DSEV----DRLLDGLNPQQRQAVVHEGSPLLIVAGAG
MAB_1054|M.abscessus_ATCC_1997 -MANLSEPALFDMPSDSSASSDALLDGLNPQQRAAVAHQGAPLLIVAGAG
TH_1135|M.thermoresistible__bu MMVAMPAEAS----SLR----ARLLDDLDDEQREAVTAPRGPVCVLAGAG
* *::.*: :** ** *: ::****
Mflv_1861|M.gilvum_PYR-GCK SGKTAVLTRRIAYLLAARDVGVGQVLAITFTNKAAAEMRERVVGLVGPRA
Mvan_4873|M.vanbaalenii_PYR-1 SGKTAVLTRRIAYLLAARDVGVGQVLAITFTNKAAAEMRERVVSLVGPRA
MSMEG_5534|M.smegmatis_MC2_155 SGKTAVLTRRIAYLLAEREVGVGQVLAITFTNKAAAEMRERVVQLVGPRA
MMAR_4553|M.marinum_M SGKTAVLTRRIAYLIAARGVGVGQILAITFTNKAAAEMRERVVSLVGGRA
MUL_4723|M.ulcerans_Agy99 SGKTAVLTRRIAYLIAARGVGVGQILAITFTNKAAAEVRERVVSLVGGRA
Mb0974|M.bovis_AF2122/97 SGKTAVLTRRIAYLMAARGVGVGQILAITFTNKAAAEMRERVVGLVGEKA
Rv0949|M.tuberculosis_H37Rv SGKTAVLTRRIAYLMAARGVGVGQILAITFTNKAAAEMRERVVGLVGEKA
MAV_1071|M.avium_104 SGKTAVLTRRVAYLIAARGVGVGQVLAITFTNKAAAEMRERVVRLVGNRA
MLBr_00153|M.leprae_Br4923 SGKTTVLARRIAYLIAARSVGVSQILAITFTNKAAAEMRERVARLVGDHT
MAB_1054|M.abscessus_ATCC_1997 SGKTAVLTRRIAYLLAARDVSPGQILAITFTNKAAAEMRERVGRIVANRV
TH_1135|M.thermoresistible__bu TGKTRTITRRIAYLVAAGHVAPSQVLAVTFTQRAAGEMRGRLRALDDGVG
:*** .::**:***:* *. .*:**:***::**.*:* *: :
Mflv_1861|M.gilvum_PYR-GCK R-NMWVSTFHSTCVRILRNQASLLPGLNSNFSIYDADDSRRLLLMIGKDM
Mvan_4873|M.vanbaalenii_PYR-1 R-NMWVSTFHSTCVRILRNQASLLPGLNSNFSIYDADDSRRLLLMIGKDM
MSMEG_5534|M.smegmatis_MC2_155 K-AMWVSTFHSTCVRILRNQASLLPGLNSNFSIYDSDDSRRLLMMIGKDL
MMAR_4553|M.marinum_M K-YMWVSTFHSTCVRILRNQAALIKGLNSNFSIYDADDSRRLLQMIGRDM
MUL_4723|M.ulcerans_Agy99 K-YMWVSTFHSTCVRILRNQAALIKGLNSNFSIYDADDSRRLLQMIGRDM
Mb0974|M.bovis_AF2122/97 R-YMWVSTFHSTCVRILRNQAALIEGLNSNFSIYDADDSRRLLQMVGRDL
Rv0949|M.tuberculosis_H37Rv R-YMWVSTFHSTCVRILRNQAALIEGLNSNFSIYDADDSRRLLQMVGRDL
MAV_1071|M.avium_104 R-AMWVSTFHSTCVRILRNQASLIEGLNSNFSIYDADDSRRLLQMIGRDM
MLBr_00153|M.leprae_Br4923 GPSMWVSTFHSTCVRILRNQASLIGGLNSNFSIYDVDDSRSLLQMIGQDM
MAB_1054|M.abscessus_ATCC_1997 Q-SMWVSTFHSSCVRILRNQASLLPGLNSNFSIYDSDDSRRLLQMIGRDM
TH_1135|M.thermoresistible__bu TGAVQAMTFHAAARRQLR---YFWPQVVGDAPWQLLDSKFSVVASAANRC
: . ***::. * ** : : .: . *.. :: ..
Mflv_1861|M.gilvum_PYR-GCK GLDTKRHSPRLLANSISNLKNELIGPEQAAAEASEAADD------LARIV
Mvan_4873|M.vanbaalenii_PYR-1 GLDTKRHSPRLLANGISNLKNELIGPEQAAAEASEAEDD------LARII
MSMEG_5534|M.smegmatis_MC2_155 GLDTKRYSPRLLANGISNLKNELIGPEQAAAEASEASDE------MATVI
MMAR_4553|M.marinum_M GLDIKRYSPRLLANAISNLKNELIDPDRAVSDLSEDSDD------LARTV
MUL_4723|M.ulcerans_Agy99 GLDIKRYSPRLLTNAISNLKNELIDPDRAVSDLSEDSDD------LARTV
Mb0974|M.bovis_AF2122/97 GLDIKRYSPRLLANAISNLKNELIDPHQALAGLTEDSDD------LARAV
Rv0949|M.tuberculosis_H37Rv GLDIKRYSPRLLANAISNLKNELIDPHQALAGLTEDSDD------LARAV
MAV_1071|M.avium_104 GLDIKRYSPRLLANAISNLKNELIDPAAAVANLTDDSDE------LSRTV
MLBr_00153|M.leprae_Br4923 GLDIKRYSPRLMANAISNLKNELIDAESAVANLSSDTDD------LARTV
MAB_1054|M.abscessus_ATCC_1997 GLDIKRYSPRLLATAISNLKNELISPEQAVANLTAGGQDGELRGDLPQIV
TH_1135|M.thermoresistible__bu GLPTGTDDVRDLAGEIEWAKASLITPEDYPKAVAEAGRDIP---FDAAKV
** . * :: *. * .** . : : . :
Mflv_1861|M.gilvum_PYR-GCK ADVYG--EYQRRLRAANALDFDDLIGETVAVLQAFPQIAQYYRRRFRHIL
Mvan_4873|M.vanbaalenii_PYR-1 ASVYA--EYQRRLRAANALDFDDLIGETVAVLQAFPQIAQYYRRRFRHIL
MSMEG_5534|M.smegmatis_MC2_155 AQVYG--EYQRRLRAANALDFDDLIGETVAVLQAFPQIAQYYRRRFRHIL
MMAR_4553|M.marinum_M ASVYA--EYQRRLRAANALDFDDLIGETVGILQAFPDIAGHYRRRFRHVL
MUL_4723|M.ulcerans_Agy99 ASVYA--EYQRRLRAANALDFDDLIGETVGILQAFPDIAGHYRRRFRHVL
Mb0974|M.bovis_AF2122/97 ASVYD--EYQRRLRAANALDFDDLIGETVAVLQAFPQIAQYYRRRFRHVL
Rv0949|M.tuberculosis_H37Rv ASVYD--EYQRRLRAANALDFDDLIGETVAVLQAFPQIAQYYRRRFRHVL
MAV_1071|M.avium_104 ASVYG--EYQRRLRAANALDFDDLIGETVAVLRAFPQIAQHYRRRFRHVL
MLBr_00153|M.leprae_Br4923 ATVYG--EYQRRLRTANALDFDDLIGETVGILQAFPQIAEHYRRRFRHVL
MAB_1054|M.abscessus_ATCC_1997 ADVYG--EYQRRLRAANALDFDDLIGETVAVLQQFPQIAQFYRRKFRHIL
TH_1135|M.thermoresistible__bu AAAYAGYEALKARQETVLLDFDDLLLHTAAAIENEPAVAQEFRDRYRCFV
* .* * : : : ******: .*.. :. * :* :* ::* .:
Mflv_1861|M.gilvum_PYR-GCK VDEYQDTNHAQYVLVRELTGSGATESDE--SVAPAELCVVGDADQSIYAF
Mvan_4873|M.vanbaalenii_PYR-1 VDEYQDTNHAQYVLVRELVG---TETVD--GVPPAELCVVGDADQSIYAF
MSMEG_5534|M.smegmatis_MC2_155 VDEYQDTNHAQYVLVRELVG-HHLEDHD--GVPPSELCVVGDADQSIYAF
MMAR_4553|M.marinum_M VDEYQDTNHAQYVLVRELVGRGSEDEPG--DVPPAQLCVVGDADQSIYAF
MUL_4723|M.ulcerans_Agy99 VDEYQDTNHAQYVLVRELVGRGSEDEPG--DVPPAQLCVVGDADQSIYAF
Mb0974|M.bovis_AF2122/97 VDEYQDTNHAQYVLVRELVGRDSND-----GIPPGELCVVGDADQSIYAF
Rv0949|M.tuberculosis_H37Rv VDEYQDTNHAQYVLVRELVGRDSND-----GIPPGELCVVGDADQSIYAF
MAV_1071|M.avium_104 VDEYQDTNHAQYVLVRELVGTDTED-----GVPPGELCVVGDADQSIYAF
MLBr_00153|M.leprae_Br4923 VDEYQDTNHAQYVLVRELVGHGARNSPD--DMPPGELCVVGDADQSIYAF
MAB_1054|M.abscessus_ATCC_1997 VDEYQDTNHAQYVLVRELVGAHVDRTVDPDGPEPAELCVVGDADQSIYAF
TH_1135|M.thermoresistible__bu VDEYQDVTPLQQRVLNAWLG------------ERDDLTVVGDANQTIYSF
******.. * ::. * :* *****:*:**:*
Mflv_1861|M.gilvum_PYR-GCK RGATIRNIEDFERDFPSATTILLEQNYRSTQTILNAANSVIARNAGRREK
Mvan_4873|M.vanbaalenii_PYR-1 RGATIRNIEDFERDFPNATTILLEQNYRSTQTILNAANSVIARNVGRRDK
MSMEG_5534|M.smegmatis_MC2_155 RGATIRNIEDFERDFPDATTILLEQNYRSTQTILNAANAVIARNTGRREK
MMAR_4553|M.marinum_M RGATIRNIEDFERDYPDATTILLEQNYRSTQNILSAANSVISRNSGRREK
MUL_4723|M.ulcerans_Agy99 RGATIRNIEDFERDYPDATTILLEQNYRSTQNILSAANSVISRNSGRREK
Mb0974|M.bovis_AF2122/97 RGATIRNIEDFERDYPDTRTILLEQNYRSTQNILSAANSVIARNAGRREK
Rv0949|M.tuberculosis_H37Rv RGATIRNIEDFERDYPDTRTILLEQNYRSTQNILSAANSVIARNAGRREK
MAV_1071|M.avium_104 RGATIRNIEDFERDYPDATTILLEQNYRSTQNILSAANSVIARNSGRRDK
MLBr_00153|M.leprae_Br4923 RGSTISNIEDFERDYPDTTTILLEQNYRSTQNILSAANSVIARNFGRRDK
MAB_1054|M.abscessus_ATCC_1997 RGATIRNIEEFERDYPNAKTILLEQNYRSTQNILSAANSVIAKNANRREK
TH_1135|M.thermoresistible__bu TGATPQYLLDFSRRFPDATVVRLERDYRSTPQVVSLANRVIATARGRMAG
*:* : :*.* :*.: .: **::**** ::. ** **: .*
Mflv_1861|M.gilvum_PYR-GCK RLWTDAGE---GELIVGYVADNEHDEARFIAQEIDALSDSIPDFSYNDVA
Mvan_4873|M.vanbaalenii_PYR-1 RLWTDAGE---GELIVGYVADNEHDEARFVAQEIDALSDAVADFSYNDVA
MSMEG_5534|M.smegmatis_MC2_155 RLWTDAGQ---GELIVGYVADNEHDEARFVADEIDALTDRHG-YNYNDIA
MMAR_4553|M.marinum_M RLWTDAGA---GELIVGYVADNEHDEARFVAEEIDALAERDE-ITYNDVA
MUL_4723|M.ulcerans_Agy99 RLWTDAGA---GELIVGYVADNEHDEARFVAEEIDALAERDE-ITYSDVA
Mb0974|M.bovis_AF2122/97 RLWTDAGA---GELIVGYVADNEHDEARFVAEEIDALAEGSE-ITYNDVA
Rv0949|M.tuberculosis_H37Rv RLWTDAGA---GELIVGYVADNEHDEARFVAEEIDALAEGSE-ITYNDVA
MAV_1071|M.avium_104 RLWTDAGA---GELIVGYVADNEHDEARFVAEEIDALAQKGE-ITYNDVA
MLBr_00153|M.leprae_Br4923 RLWTDAGK---GELIVGYVSDNEHDEAKFIADEIDALVGGGE-ITYDDVA
MAB_1054|M.abscessus_ATCC_1997 RLWTDSGE---GELIVGYVADNEHDEASFIAREIDGLFDRSD-ANYGDIA
TH_1135|M.thermoresistible__bu SKLHLVGQRPPGPQPTFSEYPDEVAEATAVAKNIARLIESGV--APAEIA
* * . :* ** :* :* * ::*
Mflv_1861|M.gilvum_PYR-GCK VFYRTNNSSRAIEEVFIRAGIPYKVVGGVRFYERREIRDIVAYLRVLDNP
Mvan_4873|M.vanbaalenii_PYR-1 VFYRTNNSSRAIEEVFIRAGIPYKVVGGVRFYERREIRDIVAYLRVLDNP
MSMEG_5534|M.smegmatis_MC2_155 VFYRTNNSSRALEEVFIRAGIPYKVVGGVRFYERREIRDIVAYLRVLDNP
MMAR_4553|M.marinum_M VFYRTNNSSRSLEEVFIRAGIPYKVVGGVRFYERKEIRDIIAYLRVLDNP
MUL_4723|M.ulcerans_Agy99 VFYRTNNSSRSLEEVFIRAGIPYKVVGGVRFYERKEIRDIVAYLRVLDNP
Mb0974|M.bovis_AF2122/97 VFYRTNNSSRSLEEVLIRAGIPYKVVGGVRFYERKEIRDIVAYLRVLDNP
Rv0949|M.tuberculosis_H37Rv VFYRTNNSSRSLEEVLIRAGIPYKVVGGVRFYERKEIRDIVAYLRVLDNP
MAV_1071|M.avium_104 VFYRTNNSSRSFEEVFIRAGIPYKVVGGVRFYERKEIRDIIAYLRVLDNP
MLBr_00153|M.leprae_Br4923 VLYRANNLSRSLEEVFIPTGIPYKVVGGFCFYESKEIRDLIAYLRVLDNP
MAB_1054|M.abscessus_ATCC_1997 VFYRTNNNSRSLEEIFIRTGIPYKVVGGVRFYERKEIRDIVAYLRVLTNP
TH_1135|M.thermoresistible__bu VLYRINAQSEVYEEALTEAGIAFQVRGGEGFFSRQEIRQALLVLQRA---
*:** * *. ** : :**.::* ** *:. :***: : *:
Mflv_1861|M.gilvum_PYR-GCK GDAVSMRRILNTPRRGIGDRAEACVAVHAENTGLNFNDALVAAAEGNVPM
Mvan_4873|M.vanbaalenii_PYR-1 GDSVSMRRILNTPRRGIGDRAEACVAVYAETTGANFNDALAAAAEGKVPM
MSMEG_5534|M.smegmatis_MC2_155 GDSVSMRRILNTPRRGIGDRAEACVAVYAENTGVSFNEALQAAAEGRVPM
MMAR_4553|M.marinum_M GDAVSMRRILNTPRRGIGDRAEACVAVYAENTGAGFSDALVAAAEGKVPM
MUL_4723|M.ulcerans_Agy99 GDAVSMRRILNTPRRGIGDRAEACVAVYAENTGAGFSDALVAAAEGKVPM
Mb0974|M.bovis_AF2122/97 GDAVSLRRILNTPRRGIGDRAEACVAVYAENTGVGFGDALVAAAQGKVPM
Rv0949|M.tuberculosis_H37Rv GDAVSLRRILNTPRRGIGDRAEACVAVYAENTGVGFGDALVAAAQGKVPM
MAV_1071|M.avium_104 GDAVSMRRILNTPRRGIGDRAEACVAVYAENTGASFADALVAAAEGKVPM
MLBr_00153|M.leprae_Br4923 GDAVSMRRILNTPRRGIGDRAEACVSVYAENTGTSFADALQAVAEGKVPM
MAB_1054|M.abscessus_ATCC_1997 GDAVSLRRILNTPRRGIGDRAEACVAVHAETNGIGFGEALADAAAGKVAM
TH_1135|M.thermoresistible__bu ----------------------------AERAGTGAVETPAG--------
** * . ::
Mflv_1861|M.gilvum_PYR-GCK LNTRSEKAIAGFVALLDDLRGGLDG-------ELGDLVEAVLDRTGYRAE
Mvan_4873|M.vanbaalenii_PYR-1 LNTRSEKAIKGFVELLDELRGGLDG-------ELGDLVETVLDRTGYRAE
MSMEG_5534|M.smegmatis_MC2_155 LNTRSEKCIASFVQMLDDLRGKLDE-------ELGDLVEAVLERTGYRRE
MMAR_4553|M.marinum_M LNSRAEKAIAGFVELLDELRGRLDSSEG----DLGELVEAVLERTGYRRE
MUL_4723|M.ulcerans_Agy99 LNSRAEKAIAGFVELLDELRGRLDSSEG----DVGELVEAVLERTGYRRE
Mb0974|M.bovis_AF2122/97 LNTRAEKAIAGFVEMFDELRGRLDD-------DLGELVEAVLERTGYRRE
Rv0949|M.tuberculosis_H37Rv LNTRAEKAIAGFVEMFDELRGRLDD-------DLGELVEAVLERTGYRRE
MAV_1071|M.avium_104 LNSRAEKAIAGFVELLDDLRGRLDD-------DLGDLVESVLERTGYRRE
MLBr_00153|M.leprae_Br4923 LNTRSVKAIAGFVALLDDLRCRVDD-------DLGELVESVLERSGYLRE
MAB_1054|M.abscessus_ATCC_1997 LNSRSEKAIAAFMDMLDQIRVTLRTDDPGALPDIGDVVEAVLNHTGYRYE
TH_1135|M.thermoresistible__bu -------------------------------AELAAAVRGLLEPLGLTAE
::. *. :*: * *
Mflv_1861|M.gilvum_PYR-GCK LEASSDPQDLARLDNLNELVSVAHEFSTDLANARALAEGD-EDAQDEDVP
Mvan_4873|M.vanbaalenii_PYR-1 LEASSDPQDLARLDNLNELVSVAHEFSIDLANAKALASEEGAEPQDEDVP
MSMEG_5534|M.smegmatis_MC2_155 LEASSDPQDLARLDNLNELVSVAHEFSIDLANAQALAEES-DEPVDEDIP
MMAR_4553|M.marinum_M LESSTDPQELARLDNLNELVSVAHEFSTDLANAAEPEP------DAEDVP
MUL_4723|M.ulcerans_Agy99 LESSTDPQELARLDNLNELVSVAHEFSTDLANAAEPEP------DAEDVP
Mb0974|M.bovis_AF2122/97 LEASTDPQELARLDNLNELVSVAHEFSTDRENAAALGP------DDEDVP
Rv0949|M.tuberculosis_H37Rv LEASTDPQELARLDNLNELVSVAHEFSTDRENAAALGP------DDEDVP
MAV_1071|M.avium_104 LESSTDPQELARLDNLNELVSVAHEFSTDRANAAALDESL-DAADDEDVP
MLBr_00153|M.leprae_Br4923 LESSTDPQELARLDNLNELVSFAHEFSTEQANAAALAKSL-HTPEDEDVP
MAB_1054|M.abscessus_ATCC_1997 LENSSDPQELARLDNLNELVSVAHEFSADAANAQAAGEDV--PGDDEDVG
TH_1135|M.thermoresistible__bu PPAGTRAR--ERWEALAALADLVDEEVATQPGLT----------------
.: .: * : * *.....* .
Mflv_1861|M.gilvum_PYR-GCK DT-GVLAAFLERVSLVADADDIPEHGSGVVTMMTLHTAKGLEFPLVFVTG
Mvan_4873|M.vanbaalenii_PYR-1 DT-GVLAAFLERVSLVADADDIPEHGAGVVTMMTLHTAKGLEFPVVFVTG
MSMEG_5534|M.smegmatis_MC2_155 DT-GVLAQFLERVSLVADADDIPEEGSGVVTMMTLHTAKGLEFPAVFVTG
MMAR_4553|M.marinum_M DT-GMLAAFLERVSLVADTDDIPEHGAGVVTLMTLHTAKGLEFPVVFVTG
MUL_4723|M.ulcerans_Agy99 DT-GMLAAFLERVSLVADTDDIPEHGAGVVTLMTLHTAKGLEFPVVFVTG
Mb0974|M.bovis_AF2122/97 DT-GVLADFLERVSLVADADEIPEHGAGVVTLMTLHTAKGLEFPVVFVTG
Rv0949|M.tuberculosis_H37Rv DT-GVLADFLERVSLVADADEIPEHGAGVVTLMTLHTAKGLEFPVVFVTG
MAV_1071|M.avium_104 DT-GVLAEFLERVSLVSDSDEIPEDGAGMVTLMTLHTAKGLEFPVVFVTG
MLBr_00153|M.leprae_Br4923 DT-GALAAFLEKVSLMSDTDQIPENNSGVVTLMTLHAAKGLEFPVVFVTG
MAB_1054|M.abscessus_ATCC_1997 APPGSLEAFLERVSLVADADELPESSAGVVTMMTLHTAKGLEFPVVFVTG
TH_1135|M.thermoresistible__bu -----LPGLLNELRRRADARHPPVVQG--VTLASLHAAKGLEWDAVFLVG
* :*:.: :*: . * . **: :**:*****: **:.*
Mflv_1861|M.gilvum_PYR-GCK WEDGMFPHMRALGDPLE---LSEERRLAYVGITRARQRLYLSRAKVRSSW
Mvan_4873|M.vanbaalenii_PYR-1 WEDGMFPHMRALGDPGE---LSEERRLAYVGITRARQRLYLSRAKVRSSW
MSMEG_5534|M.smegmatis_MC2_155 WEDGMFPHMRALGDPNE---LSEERRLAYVGITRARQRLYLSRAKVRSSW
MMAR_4553|M.marinum_M WEDGMFPHMRALDDPTE---LSEERRLAYVGITRARQRLYLSRAIVRSSW
MUL_4723|M.ulcerans_Agy99 WEDGMFPHMRALDDPTE---LSEERRLAYVGITRARQRLYLSRAIVRSSW
Mb0974|M.bovis_AF2122/97 WEDGMFPHMRALDNPTE---LSEERRLAYVGITRARQRLYVSRAIVRSSW
Rv0949|M.tuberculosis_H37Rv WEDGMFPHMRALDNPTE---LSEERRLAYVGITRARQRLYVSRAIVRSSW
MAV_1071|M.avium_104 WEDGMFPHMRSLDDPVE---LSEERRLAYVGITRARQRLYLSRAIVRSSW
MLBr_00153|M.leprae_Br4923 WEDGMLPHMRTLGDPTE---LSEERRLAYVGITRARQRLYLSRAITRSSW
MAB_1054|M.abscessus_ATCC_1997 WEDGMFPHMRALGDPVE---LAEERRLAYVGITRARQRLYLSRAKVRSSW
TH_1135|M.thermoresistible__bu LADGTLPISHALAHGPESEPVEEERRLLYVGITRARVHLALSWALARAPG
** :* ::* . * : ***** ******** :* :* * .*:.
Mflv_1861|M.gilvum_PYR-GCK GQPMLNPESRFLREIPQELIDWRRTEAPASSFSAP--VGNAG----RYGT
Mvan_4873|M.vanbaalenii_PYR-1 GQPMLNPESRFLREIPQELIDWRRTEQPSS-FSAP--VGNAG----RYGA
MSMEG_5534|M.smegmatis_MC2_155 GQPMLNPESRFLREIPQELIDWRRIEAPTPAYAAPGRMSSSGGGGIGFGS
MMAR_4553|M.marinum_M GQPMLNPESRFLREIPQELLNWRRL-APKPSFSAP--VSGAG----RFGT
MUL_4723|M.ulcerans_Agy99 GQPMLNPESRFLREIPQELLNWRRL-APKPSFSAP--VSGAG----RFGT
Mb0974|M.bovis_AF2122/97 GQPMLNPESRFLREIPQELIDWRRT-APKPSFSAP--VSGAG----RFGS
Rv0949|M.tuberculosis_H37Rv GQPMLNPESRFLREIPQELIDWRRT-APKPSFSAP--VSGAG----RFGS
MAV_1071|M.avium_104 GQPMLNPESRFLREIPQELIEWRRT-APAPSFSAP--VSGAG----RFGT
MLBr_00153|M.leprae_Br4923 GQPILNPESRFLREIPPELIDWRRS-ILTDSYSTP--ASGAS----RFGR
MAB_1054|M.abscessus_ATCC_1997 GQPMMNPESRFLQEIPQELIDWRRTDPLPGRIATG--AGGYGGGGYGSGG
TH_1135|M.thermoresistible__bu GRQGRRP-SRFLNGIAPNVGSAGDRSGPTRPRRQRGATPRCRICNAVLTA
*: .* ****. *. :: .
Mflv_1861|M.gilvum_PYR-GCK PRP---SPTRPAAGK-RPLIVLEPGDRVTHDKYGLGRVEEVSGAGESAMS
Mvan_4873|M.vanbaalenii_PYR-1 PRP---SPTRPAAGK-RPLIVLEPGDRVTHDKYGLGRVEEVSGAGESAMS
MSMEG_5534|M.smegmatis_MC2_155 PRP---SPNRPGGGRNKPLMVLQPGDRVTHDKYGLGRVEEVAGTGESAMS
MMAR_4553|M.marinum_M ARS---APVRTSTSK-RPLLVLQPGDRVTHDKYGLGRVEEVSGVGESAMS
MUL_4723|M.ulcerans_Agy99 ARS---APVRTSTSK-RPLLVLQPGDRVTHDKYGLGRVEEVSGVGESAMS
Mb0974|M.bovis_AF2122/97 ARP---SPTRSGASR-RPLLVLQVGDRVTHDKYGLGRVEEVSGVGESAMS
Rv0949|M.tuberculosis_H37Rv ARP---SPTRSGASR-RPLLVLQVGDRVTHDKYGLGRVEEVSGVGESAMS
MAV_1071|M.avium_104 PRA---APTRAAMSK-RPLLVLAPGDRVTHDKYGLGRVEEVSGAGESAMS
MLBr_00153|M.leprae_Br4923 VRP---SSIRSGASK-RALLVLAPGDRVTHDKYGLGRVEEVSGVGESAIS
MAB_1054|M.abscessus_ATCC_1997 SRAGGGSLGAPSKKRTKPLMVLEAGDRVSHDKYGLGRVVEVTGVGESATS
TH_1135|M.thermoresistible__bu PPAIMLRRCESCPADIDEELLAELKEWRSRTAKEMSVPAYVVFTDNTLIA
. . :: : :: :. * ..:: :
Mflv_1861|M.gilvum_PYR-GCK LIDFGSAGRVKLMHNHAPISKL----------------
Mvan_4873|M.vanbaalenii_PYR-1 LIDFGSAGRVKLMHNHAPVSKL----------------
MSMEG_5534|M.smegmatis_MC2_155 LIDFGSAGRVKLMHNHAPLQKL----------------
MMAR_4553|M.marinum_M LIDFGSSGRVKLMHNHAPVAKL----------------
MUL_4723|M.ulcerans_Agy99 LIDFGSSGRVKLMHNHAPVAKL----------------
Mb0974|M.bovis_AF2122/97 LIDFGSSGRVKLMHNHAPVTKL----------------
Rv0949|M.tuberculosis_H37Rv LIDFGSSGRVKLMHNHAPVTKL----------------
MAV_1071|M.avium_104 LIDFGSSGRVKLMHNHAPISKL----------------
MLBr_00153|M.leprae_Br4923 LIDFGSSGRIKLMHNHAPVTKL----------------
MAB_1054|M.abscessus_ATCC_1997 LIDFGSEGRVKLMHNFAPVQKL----------------
TH_1135|M.thermoresistible__bu IAEMLPTDDAALVAIPGIGARKLEQFGPDVLAMVKNRR
: :: . . *: . :