For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNAPRAWPAEPTATPRVKLTNADKVLYPATGTTKADVFDYYTRIADVMIPHIAGRPVTRKRWPNGVDQES FFEKQLASSAPDWLPRASVTHRSGTTTYPIIDSATGLAWIAQQAALEVHVPQWRFVAEWTRSRAEELKPG PATRLVFDLDPGEGVTMAQLAEVARAVRDLIDGIGLQTFPLTSGSKGLHLYTPLTEPVSSKGATVLAKRV AQQLEKTMPKLVTSTMTKSLRAGKIFVDWSQNNGSKTTIAPYSLRGRDHPTVAAPRTWDELDDPALRHLR YDEVLTRVARDGDLLAELDASDNQTPVPDRLTKYRSMRDASKTPEPVPAAKPAAGRGNTFVIQEHHARRL HYDFRLERDGVLVSWAVPKNLPDTPAVNHLAVHTEDHPLEYGGFEGVIPKGEYGAGRVVIWDSGTYDAEK FQDDEVIVNLHGRKISGRYALIQTEGDQWLAHRMKDQNVFDFDTIAPMLATHGSVSALKASQWAFEGKWD GYRLLVEADHGTLRVRSRRGREVTGEYRELRSLAKALEEHHAVLDGEAVVLDKRGVPNFHEMQNRGKSAR VEFWAFDLLYLDGRSLLRARYRDRRKLLEMLASGGDLTVPELLPGDGDQALRQSAERGWEGVIAKRRDST YQPGRRSSSWIKDKHWNTQEVVIGGWKAGEGGRSSGIGSLLMGIPGPGGLHFAGRVGTGFTERDLANLKK TLAPLRTDQSPFDAPLPRSEAKGVTFVEPVLVGEVRYSEWTPDDRLRQSSWRGLRPDKEASEVVRE
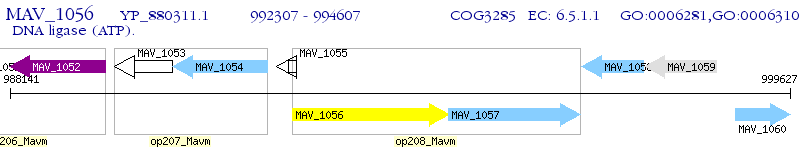
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1056 | - | - | 100% (766) | ATP-dependent DNA ligase |
| M. avium 104 | MAV_3148 | ligD | 7e-43 | 37.97% (295) | DNA polymerase LigD ligase subunit |
| M. avium 104 | MAV_4893 | - | 2e-36 | 31.08% (296) | hypothetical protein MAV_4893 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0963 | - | 0.0 | 79.29% (758) | ATP-dependent DNA ligase |
| M. gilvum PYR-GCK | Mflv_1828 | - | 0.0 | 71.01% (759) | ATP-dependent DNA ligase |
| M. tuberculosis H37Rv | Rv0938 | - | 0.0 | 79.16% (758) | ATP-dependent DNA ligase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1033 | - | 0.0 | 67.55% (758) | ATP-dependent DNA ligase |
| M. marinum M | MMAR_4573 | - | 0.0 | 81.40% (774) | ATP dependent DNA ligase |
| M. smegmatis MC2 155 | MSMEG_5570 | - | 0.0 | 70.87% (762) | ATP-dependent DNA ligase |
| M. thermoresistible (build 8) | TH_3391 | - | 0.0 | 69.91% (761) | PUTATIVE ATP dependent DNA ligase |
| M. ulcerans Agy99 | MUL_4434 | - | 0.0 | 81.14% (774) | ATP-dependent DNA ligase |
| M. vanbaalenii PYR-1 | Mvan_4915 | - | 0.0 | 70.87% (769) | ATP-dependent DNA ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1828|M.gilvum_PYR-GCK MVDRYDR------------VKLTNPDKVLYPASG-----TTKAQVFEYYV
Mvan_4915|M.vanbaalenii_PYR-1 MVDRYER------------VKLTNPDKVLYPATGDRRRAVTKAEVFDYYV
MSMEG_5570|M.smegmatis_MC2_155 -MERYER------------VRLTNPDKVLYPATG-----TTKAEVFDYYL
TH_3391|M.thermoresistible__bu -MERYER------------VKLTNPDKVLYPASG-----TTKAEVFEYYI
MMAR_4573|M.marinum_M ---MGSAQAWPPGQANGPQVTLTNADKVLYPATG-----TTKSDVFGYYT
MUL_4434|M.ulcerans_Agy99 ---MGSAQAWPPGQANGPQVTLTNADKVLYPATG-----TTKSDVFGYYT
Mb0963|M.bovis_AF2122/97 ---MGSASEQ--------RVTLTNADKVLYPATG-----TTKSDIFDYYA
Rv0938|M.tuberculosis_H37Rv ---MGSASEQ--------RVTLTNADKVLYPATG-----TTKSDIFDYYA
MAV_1056|M.avium_104 ---MNAPRAWPAEPTATPRVKLTNADKVLYPATG-----TTKADVFDYYT
MAB_1033|M.abscessus_ATCC_1997 ---------------------------MLYPATDTSP-ATTKADVFGYYT
:****:. .**:::* **
Mflv_1828|M.gilvum_PYR-GCK NVAEGMLPHIAGRAVTRKRWPNGVEESSFFEKQLASSAPDWLDRGTIAHK
Mvan_4915|M.vanbaalenii_PYR-1 NVAAAMLPHIAGRAVTRKRWPNGVTEESFFEKQLASSAPDWLGRGTIVHK
MSMEG_5570|M.smegmatis_MC2_155 SIAQVMVPHIAGRPVTRKRWPNGVAEEAFFEKQLASSAPSWLERGSITHK
TH_3391|M.thermoresistible__bu EIADAMLPHIAGRPVTRKRWPNGVHEASFFEKQLASSAPDWLARGTITHK
MMAR_4573|M.marinum_M GIAEVMIPHIAGRPATRKRWPNGVDQASFFEKQLASSAPDWLPRASVVHR
MUL_4434|M.ulcerans_Agy99 GIAEVMIPHIAGRPATRKRWPNGVDQASFFEKQLASSAPDWLPRASVVHR
Mb0963|M.bovis_AF2122/97 GVAEVMLGHIAGRPATRKRWPNGVDQPAFFEKQLALSAPPWLSRATVAHR
Rv0938|M.tuberculosis_H37Rv GVAEVMLGHIAGRPATRKRWPNGVDQPAFFEKQLALSAPPWLSRATVAHR
MAV_1056|M.avium_104 RIADVMIPHIAGRPVTRKRWPNGVDQESFFEKQLASSAPDWLPRASVTHR
MAB_1033|M.abscessus_ATCC_1997 AIAPFMLPHIAGRPVTRKRWPNGIDQPSFFEKDLASSAPDWLPRRRIEHK
:* *: *****..********: : :****:** *** ** * : *:
Mflv_1828|M.gilvum_PYR-GCK SGTTTYPVIDTVEGLAWIAQQAALEVHVPQWRFTRGGQSGAADGGDGKPG
Mvan_4915|M.vanbaalenii_PYR-1 SGTTTYPVIDTVEGLAWIAQQAALEVHVPQWRFV-----------DGSVG
MSMEG_5570|M.smegmatis_MC2_155 SGTTTYPIINTREGLAWVAQQASLEVHVPQWRFEDGDQ-----------G
TH_3391|M.thermoresistible__bu SRTTTYPIIDTREGLAWIAQQAALEVHVPQWRFVAGGRSGR--DGKLTMG
MMAR_4573|M.marinum_M SGTTTYPIIDSATGLAWIAQQAALEVHVPQWRFVAEWT---SKGEILTPG
MUL_4434|M.ulcerans_Agy99 SGTTTYPIIDSATGLAWIAQQAALEVHVPQWRFVAEWT---SKGEILTPG
Mb0963|M.bovis_AF2122/97 SGTTTYPIIDSATGLAWIAQQAALEVHVPQWRFVAEPG---SGE--LNPG
Rv0938|M.tuberculosis_H37Rv SGTTTYPIIDSATGLAWIAQQAALEVHVPQWRFVAEPG---SGE--LNPG
MAV_1056|M.avium_104 SGTTTYPIIDSATGLAWIAQQAALEVHVPQWRFVAEWTR--SRAEELKPG
MAB_1033|M.abscessus_ATCC_1997 SRYVSYPLIDTSVALAWIAQQAALEVHVPQWRFAGENP-----------G
* .:**:*:: .***:****:********** *
Mflv_1828|M.gilvum_PYR-GCK PATRIVFDLDPGEGVTFRQLCEVAHEVRELITDIGLTTFPLTSGSKGLHL
Mvan_4915|M.vanbaalenii_PYR-1 PATRIVFDLDPGEGVTFRQLCEVANEVRDLLADIGLRTFPLTSGSKGLHL
MSMEG_5570|M.smegmatis_MC2_155 PATRIVFDLDPGEGVTMTQLCEIAHEVRALMTDLDLETYPLTSGSKGLHL
TH_3391|M.thermoresistible__bu PATRIVFDLDPGEGVTMRELCRVAHEVRDLITDIGLDTFPLTSGSKGLHL
MMAR_4573|M.marinum_M PATRLVFDLDPGEGVTMAQLAQVARAVRDLLADIGLSTLPLTSGSKGLHL
MUL_4434|M.ulcerans_Agy99 PATRLVFDLDPGEGVTMAQLAQVARAVRDLLADIGLSTLPLTSGSKGLHF
Mb0963|M.bovis_AF2122/97 PATRLVFDLDPGEGVMMAQLAEVARAVRDLLADIGLVTFPVTSGSKGLHL
Rv0938|M.tuberculosis_H37Rv PATRLVFDLDPGEGVMMAQLAEVARAVRDLLADIGLVTFPVTSGSKGLHL
MAV_1056|M.avium_104 PATRLVFDLDPGEGVTMAQLAEVARAVRDLIDGIGLQTFPLTSGSKGLHL
MAB_1033|M.abscessus_ATCC_1997 PATRLVFDLDPGEGVGMAQLAEVARAIRDVLADIGLTTFPVTSGSKGLHL
****:********** : :*..:*. :* :: .:.* * *:********:
Mflv_1828|M.gilvum_PYR-GCK YVPLEDPISSRGASVLAKRVAQQLEQSMPKRVTATMTKSLRDGKVFLDWS
Mvan_4915|M.vanbaalenii_PYR-1 YVPLDEPISSRGASVLAKRVAQQLEQAMPMQVTATMTKSLRTGKVFVDWS
MSMEG_5570|M.smegmatis_MC2_155 YVPLAEPISSRGASVLARRVAQQLEQAMPKLVTATMTKSLRAGKVFLDWS
TH_3391|M.thermoresistible__bu YVPLDEPISSQGASVLAKRVALQLEKAMPTLVTATMTKSVRTGKVFLDWS
MMAR_4573|M.marinum_M YTPLDEPVSSKGATVLAKRVAQQLETVMPALVTATMTKSLRAGKVFLDWS
MUL_4434|M.ulcerans_Agy99 YTPLDEPVSSKGATVLAKRVAQQLETVMPALVTATMTKSLRAGKVFLDWS
Mb0963|M.bovis_AF2122/97 YTPLDEPVSSRGATVLAKRVAQRLEQAMPALVTSTMTKSLRAGKVFVDWS
Rv0938|M.tuberculosis_H37Rv YTPLDEPVSSRGATVLAKRVAQRLEQAMPALVTSTMTKSLRAGKVFVDWS
MAV_1056|M.avium_104 YTPLTEPVSSKGATVLAKRVAQQLEKTMPKLVTSTMTKSLRAGKIFVDWS
MAB_1033|M.abscessus_ATCC_1997 YVPLAQPASSSGAVAVAKKVAVQLEQVMPDLVTATMTRQLRAGKVFVDWS
*.** :* ** ** .:*::** :** ** **:***:.:* **:*:***
Mflv_1828|M.gilvum_PYR-GCK QNNGNKTTIAPYSLRGRERPTVAAPRTWDEIEDPDLRHLTFDEVLDRLER
Mvan_4915|M.vanbaalenii_PYR-1 QNNANKTTIAPYSLRGREHPTVAAPRTWEEISDPDLRQLTFDEVLDRLDR
MSMEG_5570|M.smegmatis_MC2_155 QNNAAKTTIAPYSLRGRDHPTVAAPRTWDEIADPELRHLRFDEVLDRLDE
TH_3391|M.thermoresistible__bu QNNAAKTTIAPYSLRGREHPTVAAPRTWDEIDDPDLRHLTFTEVLDRVDR
MMAR_4573|M.marinum_M QNSGSKTTIAPYSLRGREQPTVAAPRSWEELDDPGLRQLTYDEVLARLTR
MUL_4434|M.ulcerans_Agy99 QNSGSKTTIAPYSLRGREQPTVAAPRSWEELDDPGLRQLTYDEVLARLTR
Mb0963|M.bovis_AF2122/97 QNSGSKTTIAPYSLRGRTHPTVAAPRTWAELDDPALRQLSYDEVLTRIAR
Rv0938|M.tuberculosis_H37Rv QNSGSKTTIAPYSLRGRTHPTVAAPRTWAELDDPALRQLSYDEVLTRIAR
MAV_1056|M.avium_104 QNNGSKTTIAPYSLRGRDHPTVAAPRTWDELDDPALRHLRYDEVLTRVAR
MAB_1033|M.abscessus_ATCC_1997 QNSGSKTTVAPYSLRGRAVPTVAAPRTWEELDDPGLRQLRYDEVLTRAQR
**.. ***:******** *******:* *: ** **:* : *** * .
Mflv_1828|M.gilvum_PYR-GCK DGDLLADLDPP---LPATDKLTRYRSMRDRTKTPEPVPAEPPETGANDKF
Mvan_4915|M.vanbaalenii_PYR-1 DGDLLAGLDPP---VPVPDRLTRYRSMRDTAKTPEPIPGDAPTTGNNDKF
MSMEG_5570|M.smegmatis_MC2_155 YGDLLAPLDAD---APIADKLTTYRSMRDASKTPEPVPKEIPKTGNNDKF
TH_3391|M.thermoresistible__bu LGDLLAPMDAD---APVADRLSSYRSMRDPARTPEPVPKTVAATGDDNRF
MMAR_4573|M.marinum_M DGDLIAALDAD---LPGDDRLLKYRSMRNASKTPEPVPRARPVVGQGNTF
MUL_4434|M.ulcerans_Agy99 DGDLIAALDAD---LPGDDRLLKYRSMRNASKTPEPVPRARPVVGQGNTF
Mb0963|M.bovis_AF2122/97 DGDLLERLDAD---APVADRLTRYRRMRDASKTPEPIPTAKPVTGDGNTF
Rv0938|M.tuberculosis_H37Rv DGDLLERLDAD---APVADRLTRYRRMRDASKTPEPIPTAKPVTGDGNTF
MAV_1056|M.avium_104 DGDLLAELDASDNQTPVPDRLTKYRSMRDASKTPEPVPAAKPAAGRGNTF
MAB_1033|M.abscessus_ATCC_1997 DGDLLAGLDDV---EIAEDRLSTYRSMRDPTRTPEPVPAVKPEAGQDNSF
***: :* *:* ** **: ::****:* . .* .: *
Mflv_1828|M.gilvum_PYR-GCK VIQEHHARRLHYDFRLERDGVLVSWAVPKNLPDTPSVNHLAVHTEDHPMD
Mvan_4915|M.vanbaalenii_PYR-1 VIQEHHARRLHYDFRLERDGVLVSWAVPKNLPDTPSVNHLAVHTEDHPME
MSMEG_5570|M.smegmatis_MC2_155 VIQEHHARRLHYDLRLERDGVLVSFAVPKNLPETTAENRLAVHTEDHPIE
TH_3391|M.thermoresistible__bu VIQEHHARRLHYDLRLERDGVLVSWAIPKNLPDDPSVNHLAVHTEDHPLE
MMAR_4573|M.marinum_M VIQEHHARRLHYDFRLERDGVLVSWAVPKNLPETTSVNHLAVHTEDHPLE
MUL_4434|M.ulcerans_Agy99 VIQEHHARRLHDDFRLERDGVLVSWAVPKNLPETTSVNHLAVHTEDHPLE
Mb0963|M.bovis_AF2122/97 VIQEHHARRPHYDFRLERDGVLVSWAVPKNLPDNTSVNHLAIHTEDHPLE
Rv0938|M.tuberculosis_H37Rv VIQEHHARRPHYDFRLECDGVLVSWAVPKNLPDNTSVNHLAIHTEDHPLE
MAV_1056|M.avium_104 VIQEHHARRLHYDFRLERDGVLVSWAVPKNLPDTPAVNHLAVHTEDHPLE
MAB_1033|M.abscessus_ATCC_1997 VIQEHHATALHYDFRLERDGVLVSWAVPKNLPLEPSVNHLAVHTEDHPLE
******* * *:*** ******:*:***** .: *:**:******::
Mflv_1828|M.gilvum_PYR-GCK YIDFAGEIPKGEYGGGEVHIWDTGTYETEKFR---DNPPDGPAKGGEVIV
Mvan_4915|M.vanbaalenii_PYR-1 YLTFAGEIPRGEYGGGEVYIWDTGTYETEKFR---DNPPDGPDRGGEVIV
MSMEG_5570|M.smegmatis_MC2_155 YLAFHGSIPKGEYGAGDMVIWDSGSYETEKFRVP-EELDNPDDSHGEIIV
TH_3391|M.thermoresistible__bu YLTFHGEIPEGEYGGGKMLIWDTGTYEAEKFN---DVGPDEDAEGGEVII
MMAR_4573|M.marinum_M YATFEGNIPKGEYGAGKVIIWDSGTYDTEKFR----DDSRNGSEKGEVIV
MUL_4434|M.ulcerans_Agy99 YATFEGNIPKGEYGAGKVIIWDSGTYDTEKFR----DDSRNGAEKGEVIV
Mb0963|M.bovis_AF2122/97 YATFEGAIPSGEYGAGKVIIWDSGTYDTEKFH----DDPHTG----EVIV
Rv0938|M.tuberculosis_H37Rv YATFEGAIPSGEYGAGKVIIWDSGTYDTEKFH----DDPHTG----EVIV
MAV_1056|M.avium_104 YGGFEGVIPKGEYGAGRVVIWDSGTYDAEKFQ----DD--------EVIV
MAB_1033|M.abscessus_ATCC_1997 YGSFEGTIPKGEYGAGEVTIWDSGTYDTEKFIDPRDDSGADGGQKGEVIV
* * * ** ****.* : ***:*:*::*** *:*:
Mflv_1828|M.gilvum_PYR-GCK TLHGQKIEGRYALIQTDGKNWLAHRMKDQQ---KPTAADLAPMLTTEGSV
Mvan_4915|M.vanbaalenii_PYR-1 TLHGEKIDGRYALIQTDGKNWLAHRMKDQQ---RPTAADLAPMLTTHGSV
MSMEG_5570|M.smegmatis_MC2_155 TLHGEKVDGRYALIQTKGKNWLAHRMKDQK---NARPEDFAPMLATEGSV
TH_3391|M.thermoresistible__bu TLHGEKISGRYALIQTDGKNWLAHKMKDQKPRTEIRPEDFTPMLATHGSV
MMAR_4573|M.marinum_M NLHGSRISGRYALIQTNGDQWLAHRMK---DQTAFDFDTLAPMLTSHGSV
MUL_4434|M.ulcerans_Agy99 NLHGSRISGRYALIQTNGDQWLAHRMK---DQTAFDFDTLAPMLTSHGSV
Mb0963|M.bovis_AF2122/97 NLHGGRISGRYALIRTNGDRWLAHRLKNQKDQKVFEFDNLAPMLATHGTV
Rv0938|M.tuberculosis_H37Rv NLHGGRISGRYALIRTNGDRWLAHRLKNQKDQKVFEFDNLAPMLATHGTV
MAV_1056|M.avium_104 NLHGRKISGRYALIQTEGDQWLAHRMK---DQNVFDFDTIAPMLATHGSV
MAB_1033|M.abscessus_ATCC_1997 TLHGKRITGRYALIRTNGKQWLVHRMKDQRPAGSMPVG-LAPMLPTTGSV
.*** :: ******:*.*..**.*::* ::***.: *:*
Mflv_1828|M.gilvum_PYR-GCK ERLTRGQWAFEGKWDGYRLLVDADHGKLTLRSRRGRDVTAEYPQLKALAA
Mvan_4915|M.vanbaalenii_PYR-1 QRLTKGQWAFEGKWDGYRLLVEADHGRLALRSRNGRDVTAEYPQLQALAA
MSMEG_5570|M.smegmatis_MC2_155 AKYKAKQWAFEGKWDGYRVIIDADHGQLQIRSRTGREVTGEYPQFKALAA
TH_3391|M.thermoresistible__bu ARLSAETHAFEGKWDGYRLLVHIDHGKLCLRSRSGRDVTKEFPTLQAMAA
MMAR_4573|M.marinum_M AGLESGQWAFEGKWDGYRLLVEADHGALRLRSRSGRDVTAEYPQLRSLAA
MUL_4434|M.ulcerans_Agy99 AGLESGQWAFEGKWDGYRLLVEADHGALRLRSRSGRDVTTEYPQLRSLAA
Mb0963|M.bovis_AF2122/97 AGLKASQWAFEGKWDGYRLLVEADHGAVRLRSRSGRDVTAEYPQLRALAE
Rv0938|M.tuberculosis_H37Rv AGLKASQWAFEGKWDGYRLLVEADHGAVRLRSRSGRDVTAEYPQLRALAE
MAV_1056|M.avium_104 SALKASQWAFEGKWDGYRLLVEADHGTLRVRSRRGREVTGEYRELRSLAK
MAB_1033|M.abscessus_ATCC_1997 AGLTAEEWAFEGKWDGYRLILECDRGQLRALARSGRDVTAEFPALQRLAH
**********:::. *:* : :* **:** *: :: :*
Mflv_1828|M.gilvum_PYR-GCK DLADHHVILDGEAVALDEDGVPSFGEMQNRARSTRVEFWAFDILFLDGRL
Mvan_4915|M.vanbaalenii_PYR-1 DLADHHVILDGEAVALDESGVPSFREMQNRARSTRVEFWAFDILFLDGRS
MSMEG_5570|M.smegmatis_MC2_155 DLAEHHVVLDGEAVALDESGVPSFGQMQNRARSTRVEFWAFDILWLDGRS
TH_3391|M.thermoresistible__bu DLADHQVVLDGEVVALDGSGVPSFGEMQNRARATHIEFWAFDILWLDGRS
MMAR_4573|M.marinum_M DLADHHVVLDGELVALDSSGVPSFNEMQNRVRATRIEFWAFDLLFLDGRS
MUL_4434|M.ulcerans_Agy99 DLADHHVVLDGELVALDSSGVPSFNEMQNRVRATRIEFWAFDLLFLDGRS
Mb0963|M.bovis_AF2122/97 DLADHHVVLDGEAVVLDSSGVPSFSQMQNRGRDTRVEFWAFDLLYLDGRA
Rv0938|M.tuberculosis_H37Rv DLADHHVVLDGEAVVLDSSGVPSFSQMQNRGRDTRVEFWAFDLLYLDGRA
MAV_1056|M.avium_104 ALEEHHAVLDGEAVVLDKRGVPNFHEMQNRGKSARVEFWAFDLLYLDGRS
MAB_1033|M.abscessus_ATCC_1997 ELSDHRVVLDGEVVVPDRKGLPSFALLQNRTPGADIRFWAFDLLYLDGKS
* :*:.:**** *. * *:*.* :*** : :.*****:*:***:
Mflv_1828|M.gilvum_PYR-GCK LTKATYTDRRKLLEALAEG-GGLIVPPALEGD-GPEALEYARSQRWEGVI
Mvan_4915|M.vanbaalenii_PYR-1 LLRAKYTDRRRVLEALADG-GGLIVPPLIGADTGAEALETARKRGWEGVV
MSMEG_5570|M.smegmatis_MC2_155 LLRAKYSDRRKILEALADG-GGLIVPDQLPGD-GPEAMEHVRKKRFEGVV
TH_3391|M.thermoresistible__bu LLRAKHSDRRRVLLALAEK-DGLIVPDLLPGDTGAEALEYARERGFEGVV
MMAR_4573|M.marinum_M LLRARYRDRRKLLETLGSA-GNLIVPELLPGD-GAEALAYSHKRGWEGVI
MUL_4434|M.ulcerans_Agy99 LLRARYRDRRKLLETLGSA-GNLIVPELLPGD-GAEALAYSHKRGWEGVI
Mb0963|M.bovis_AF2122/97 LLGTRYQDRRKLLETLANA-TSLTVPELLPGD-GAQAFACSRKHGWEGVI
Rv0938|M.tuberculosis_H37Rv LLGTRYQDRRKLLETLANA-TSLTVPELLPGD-GAQAFACSRKHGWEGVI
MAV_1056|M.avium_104 LLRARYRDRRKLLEMLASG-GDLTVPELLPGD-GDQALRQSAERGWEGVI
MAB_1033|M.abscessus_ATCC_1997 LLRTKYRDRRRLLEVLVTGTESMTVPQQLSGD-GAAALAYSQDQGWEGVV
* : : ***::* * .: ** : .* * *: .: :***:
Mflv_1828|M.gilvum_PYR-GCK AKKRDSTYQPGRRSSAWIKDKIWNTQEVVIGGWRQGEGGRTSGIGALVLG
Mvan_4915|M.vanbaalenii_PYR-1 AKKRDSTYQPGRRSSSWIKDKLWNTQEVVIGGWRQGEGGRTSGIGALMLG
MSMEG_5570|M.smegmatis_MC2_155 AKKWDSTYQPGRRSSSWIKDKIWNTQEVVIGGWRQGEGGRSSGIGALVLG
TH_3391|M.thermoresistible__bu AKRHDSIYQPGRRSKAWIKDKIWNMQEVVIGGWRKGEGGRSSGIGALVLG
MMAR_4573|M.marinum_M AKRRDSGYQPGRRSSAWIKDKHWNTQEVVIGGWRVGEGGRSSGIGSLLMG
MUL_4434|M.ulcerans_Agy99 AKRRDSGYQPGRRSSAWIKDKHWNTQEVVIGGWRVGEGGRSSGIGSLLMG
Mb0963|M.bovis_AF2122/97 AKRRDSRYQPGRRCASWVKDKHWNTQEVVIGGWRAGEGGRSSGVGSLLMG
Rv0938|M.tuberculosis_H37Rv AKRRDSRYQPGRRCASWVKDKHWNTQEVVIGGWRAGEGGRSSGVGSLLMG
MAV_1056|M.avium_104 AKRRDSTYQPGRRSSSWIKDKHWNTQEVVIGGWKAGEGGRSSGIGSLLMG
MAB_1033|M.abscessus_ATCC_1997 AKRLDSTYVPG-RTQSWIKAKNWSSQEVVIGGWRKGQGGRSSGIGSLLMG
**: ** * ** * :*:* * *. ********: *:***:**:*:*::*
Mflv_1828|M.gilvum_PYR-GCK VPDD---DGLHFVGRVGTGFTDKQLASLKKTLEPLRTDESPFAAELPKQD
Mvan_4915|M.vanbaalenii_PYR-1 IPAGGVAGGLQFVGRVGTGFTEKELASLKETLAPLHTDESPFATELPKQD
MSMEG_5570|M.smegmatis_MC2_155 IPGP---EGLQFVGRVGTGFTEKELSKLKDMLKPLHTDESPFNAPLPKVD
TH_3391|M.thermoresistible__bu IPTE---GGLQFVGRVGTGFTDKQLAELKQLLAPLETDESPFVDPLPRPD
MMAR_4573|M.marinum_M IPGP---SGLHFAGRVGTGFTERDLANLKKTLAPLHTDESPFDAALPARE
MUL_4434|M.ulcerans_Agy99 IPGP---SGLHFAGRVGTGFTERDLANLKKTLAPLHTDESPFDAALPARE
Mb0963|M.bovis_AF2122/97 IPGP---GGLQFAGRVGTGLSERELANLKEMLAPLHTDESPFDVPLPARD
Rv0938|M.tuberculosis_H37Rv IPGP---GGLQFAGRVGTGLSERELANLKEMLAPLHTDESPFDVPLPARD
MAV_1056|M.avium_104 IPGP---GGLHFAGRVGTGFTERDLANLKKTLAPLRTDQSPFDAPLPRSE
MAB_1033|M.abscessus_ATCC_1997 IPVE---GGLRYAGRVGTGFTERELSSLKERLEPLHRAESPFDAPLSTVE
:* **::.******:::::*:.**. * **. :*** *. :
Mflv_1828|M.gilvum_PYR-GCK AKGVTFVRPELVGEVRYSERTSDGRLRQASWRGLRDDKVPEDVRWEG-
Mvan_4915|M.vanbaalenii_PYR-1 AKGVTFVRPELVGEVRYSERTSDNRLRQPSWRGLRTDKTPDEVVWE--
MSMEG_5570|M.smegmatis_MC2_155 ARGVTFVRPELVGEVRYSERTSDGRLRQPSWRGLRPDKTPDEVVWE--
TH_3391|M.thermoresistible__bu AKGVTYVRPELVGEVRYSERTADNRLRQPSWRGLRPDKSPDEVRWE--
MMAR_4573|M.marinum_M AKGVTFVAPTLVGEVRYSEWTPDDRLRQSSWRGLRPDKNPSEVVRE--
MUL_4434|M.ulcerans_Agy99 AKGVTFVVPTLVGEVRYSEWTPDDRLRQSSWRGLRPDKNPSEVVRE--
Mb0963|M.bovis_AF2122/97 AKGITYVKPALVAEVRYSEWTPEGRLRQSSWRGLRPDKKPSEVVRE--
Rv0938|M.tuberculosis_H37Rv AKGITYVKPALVAEVRYSEWTPEGRLRQSSWRGLRPDKKPSEVVRE--
MAV_1056|M.avium_104 AKGVTFVEPVLVGEVRYSEWTPDDRLRQSSWRGLRPDKEASEVVRE--
MAB_1033|M.abscessus_ATCC_1997 AKDVQFVEPVLVAEVRYGDRTPGGILKHSSWRGLRTDKSVRDVELDLG
*:.: :* * **.****.: *. . *::.****** ** :* :