For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PLESRLALRRDNLINAGVQLLGGSGGPALTVRAVCRKAALTERYFYESFTDRDEFVRAVYDDVCTRAMNT LTSATTPREAVERFVELMVDDPVRGRVLLLAPAVEPVLTRSGAEWMPNFIDLLQRKLSRIGDAVLQKMIA TSLVGGLSSLFTAYLHGQLGATRKQFIDYCTNMLYITAAPYVSAGELGKSQ*
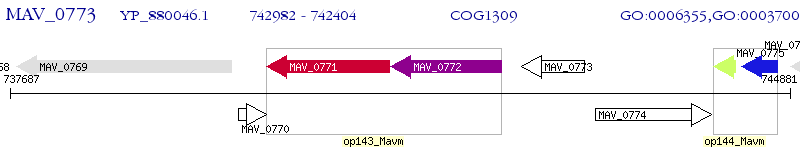
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0773 | - | - | 100% (192) | hypothetical protein MAV_0773 |
| M. avium 104 | MAV_4882 | - | 3e-21 | 34.69% (196) | FadD27 protein |
| M. avium 104 | MAV_3986 | - | 6e-19 | 31.02% (187) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0848c | - | 5e-86 | 81.77% (192) | hypothetical protein Mb0848c |
| M. gilvum PYR-GCK | Mflv_1669 | - | 4e-67 | 69.71% (175) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0825c | - | 5e-86 | 81.77% (192) | hypothetical protein Rv0825c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0756c | - | 2e-48 | 56.29% (167) | hypothetical protein MAB_0756c |
| M. marinum M | MMAR_4855 | - | 2e-81 | 77.60% (192) | hypothetical protein MMAR_4855 |
| M. smegmatis MC2 155 | MSMEG_5768 | - | 2e-71 | 74.86% (175) | hypothetical protein MSMEG_5768 |
| M. thermoresistible (build 8) | TH_1575 | - | 8e-74 | 74.73% (182) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0446 | - | 1e-81 | 77.60% (192) | hypothetical protein MUL_0446 |
| M. vanbaalenii PYR-1 | Mvan_5084 | - | 4e-68 | 69.71% (175) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0848c|M.bovis_AF2122/97 MQTGQNRGR-WSGVPLESRHALRRDNLVAAGVQLLGGAGGPALTVRAVCR
Rv0825c|M.tuberculosis_H37Rv MQTGQNRGR-WSGVPLESRHALRRDNLVAAGVQLLGGAGGPALTVRAVCR
MAV_0773|M.avium_104 -------------MPLESRLALRRDNLINAGVQLLGGSGGPALTVRAVCR
MMAR_4855|M.marinum_M MQAGQRRGR-WTGVPLEDRHAVRRDTLIAAGVQLLGEEAGPALTVRAVCR
MUL_0446|M.ulcerans_Agy99 MQAGQRRGR-WTGVPLEDRHAVRRDTLIAAGVQLLGEEAGPALTVRAVCR
Mflv_1669|M.gilvum_PYR-GCK MPSGQRRGR-WSGVPLQDRQALRRDELITAGIGLLGSSGGPNLTVRAVCK
Mvan_5084|M.vanbaalenii_PYR-1 MPSGQRRGR-WSGVPLQDRQALRRDELITAGISLLGSPGGPHLTVRAVCK
TH_1575|M.thermoresistible__bu -------------VPLSDRQTLRREELIAAGVKLLGHREGPALTVRAVCR
MSMEG_5768|M.smegmatis_MC2_155 MPSGQRRGR-WSGVPLRERTALRRDELIAAGVKLLGAADGPALTVRAVCR
MAB_0756c|M.abscessus_ATCC_199 MANGQAQRRSWAGVSPAERADERRRELMAAGVRLLGLRPRPAVTVRAVCR
:. .* ** *: **: *** * :******:
Mb0848c|M.bovis_AF2122/97 HAGLTERYFYESFADREHFVRAVYDDVCTRAMATLTSAQTPREAVEQFVE
Rv0825c|M.tuberculosis_H37Rv HAGLTERYFYESFADREHFVRAVYDDVCTRAMATLTSAQTPREAVEQFVE
MAV_0773|M.avium_104 KAALTERYFYESFTDRDEFVRAVYDDVCTRAMNTLTSATTPREAVERFVE
MMAR_4855|M.marinum_M QAALTERYFYESFSDRDHFVRAVYDDVCTRAMATLMSANTPRDAVERFVA
MUL_0446|M.ulcerans_Agy99 QAALTERYFYESFSDRDHFVRAVYDDVCTRAMATLMSANTPRDAVERFVA
Mflv_1669|M.gilvum_PYR-GCK AAGLTERYFYESFTDRDEYVAAVYDDVCTAAMATLRESESMRDAVERFVA
Mvan_5084|M.vanbaalenii_PYR-1 AAGLTERYFYESFTDRDEYVAAVYDNVCTAAMSTLMKSDSMRDAVERFVA
TH_1575|M.thermoresistible__bu EAGLTERYFYESFADRDEFVRAVYDDVCARAMSTLMTADTPRDAVERFVA
MSMEG_5768|M.smegmatis_MC2_155 EAGLTERYFYECFNDRDGFVRAVYDDVCATAMSALTAADTPRKAVEQFVS
MAB_0756c|M.abscessus_ATCC_199 SAGMTERYFYEHFSDRDTFVRAVYDHVGFRAIEALSGVRSAYEGVDAFAR
*.:******* * **: :* ****.* *: :* : ..*: *.
Mb0848c|M.bovis_AF2122/97 LMVDDPVRGRVLLLAPAVEPALTRSGAEWMPNFIELLQRKLSR-IVDPVL
Rv0825c|M.tuberculosis_H37Rv LMVDDPVRGRVLLLAPAVEPALTRSGAEWMPNFIELLQRKLSR-IVDPVL
MAV_0773|M.avium_104 LMVDDPVRGRVLLLAPAVEPVLTRSGAEWMPNFIDLLQRKLSR-IGDAVL
MMAR_4855|M.marinum_M LMVDDPVRGRVLLLAPAVEPVLTRSGADWMPNFIDLLQRKLSQ-IADPVL
MUL_0446|M.ulcerans_Agy99 LMVDDPVRGRVLLLAPAVEPVLTRSGADWMPNFIDLLQRKLSQ-IADPVL
Mflv_1669|M.gilvum_PYR-GCK LMIDDPARGRVLLLAPEAEPVLARSGARWMPNFIELLQHNLTR-ITDTTT
Mvan_5084|M.vanbaalenii_PYR-1 LMIDDPARGRVLLLAPEVEPVLARSGARWMPNFIDLLQRNLTQ-ISDPTT
TH_1575|M.thermoresistible__bu LMVDDPVRGRVLLLAPEVEPVLTRSGAKWMPNFIDLLQRKLTR-ITDPAL
MSMEG_5768|M.smegmatis_MC2_155 LMVDDPTRGRVLLLAPESEPVLTRSGAEWMPNFIDLLQRKLTR-IADPVR
MAB_0756c|M.abscessus_ATCC_199 LMVDEPTMGRVLLIAPTFEPTLSESGYDWLPRFVALLQGKLSTNLADEVQ
**:*:*. *****:** **.*:.** *:*.*: *** :*: : * .
Mb0848c|M.bovis_AF2122/97 QKLVATSLIGALTGLFTAYLNGRLGATRKQFIDYCVNMLLSTAATYAPHR
Rv0825c|M.tuberculosis_H37Rv QKLVATSLIGALTGLFTAYLNGRLGATRKQFIDYCVNMLLSTAATYAPHR
MAV_0773|M.avium_104 QKMIATSLVGGLSSLFTAYLHGQLGATRKQFIDYCTNMLYITAAPYVSAG
MMAR_4855|M.marinum_M QKLVATSLIGALTGLFTAYLNGQLGATRQQFIDYCVDLLLGTAAAYAPHR
MUL_0446|M.ulcerans_Agy99 QKLVATSLIGALTGLFTAYLNGQLGATRQQFIDYCVDLLLGTAAAYAPHR
Mflv_1669|M.gilvum_PYR-GCK QAMVATGLIGALTALFTAYLDGRLEATREQFIDHCAELLLSRAVT-----
Mvan_5084|M.vanbaalenii_PYR-1 QAMVATGLIGALTALFTAYLDGRLGATRERFIDHCAELLLSRAVT-----
TH_1575|M.thermoresistible__bu QAMVATGLIGALTALFTAYLDGRLAATREQFIDYCVGILLNPVASY----
MSMEG_5768|M.smegmatis_MC2_155 QAMVATSLIGALTALFTAYLNDRLPATREQFIDFCVDMLLGRVGSF----
MAB_0756c|M.abscessus_ATCC_199 QQLVATSLVGALTSLFTAYLNEELNVDRQRFVDYCVQILLQGANASVTPR
* ::**.*:*.*:.******. .* . *::*:*.*. :* . .
Mb0848c|M.bovis_AF2122/97 ERGESEHSIPAGPHN
Rv0825c|M.tuberculosis_H37Rv ERGESEHSIPAGPHN
MAV_0773|M.avium_104 ELGKSQ---------
MMAR_4855|M.marinum_M DRGEAERPVAARQHD
MUL_0446|M.ulcerans_Agy99 DRGEAEHPVAARQHD
Mflv_1669|M.gilvum_PYR-GCK ---------------
Mvan_5084|M.vanbaalenii_PYR-1 ---------------
TH_1575|M.thermoresistible__bu ---------------
MSMEG_5768|M.smegmatis_MC2_155 ---------------
MAB_0756c|M.abscessus_ATCC_199 PTSPSPQQPS-----