For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LQGKKIAILAADGVEKVELEQPAAALREAGAGVEVVSLQDGEIQARNHDLEPAGTFTVDRKVADASVDDF DGLVLPGGTVNPDKLRLDDTAVSFVRDFVGSGKPVAAICHGPWTLVEAGVAAGRTLTSYPSIRTDLRNAG AHVVDEEVVVDGNLITSRSPKDLPAFCSTILAQFAGATTG
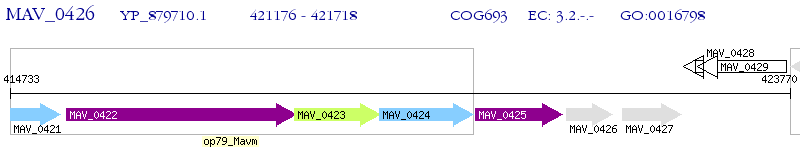
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0426 | - | - | 100% (180) | intracellular protease, PfpI family protein |
| M. avium 104 | MAV_4069 | katE | 6e-06 | 30.00% (120) | catalase HPII |
| M. avium 104 | MAV_2773 | - | 5e-05 | 27.10% (155) | DJ-1/PfpI family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1035 | - | 6e-72 | 74.14% (174) | PfpI family intracellular peptidase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02219 | purQ | 1e-05 | 34.07% (91) | phosphoribosylformylglycinamidine synthase I |
| M. abscessus ATCC 19977 | MAB_3901 | - | 7e-68 | 71.19% (177) | peptidase C56 PfpI |
| M. marinum M | MMAR_5188 | - | 6e-74 | 78.29% (175) | hypothetical protein MMAR_5188 |
| M. smegmatis MC2 155 | MSMEG_0536 | - | 1e-66 | 69.71% (175) | intracellular protease, PfpI family protein |
| M. thermoresistible (build 8) | TH_4363 | - | 2e-46 | 53.85% (182) | PUTATIVE intracellular protease, PfpI family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5796 | - | 3e-72 | 75.00% (176) | PfpI family intracellular peptidase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0426|M.avium_104 -------------MQGKKIAILAA-DGVEKVELEQPAAALREAGAGVEVV
MMAR_5188|M.marinum_M MSDE---------LNGKSIAMLAA-DGVEKVELEQPRSALLQEGARVEVL
Mflv_1035|M.gilvum_PYR-GCK MPNET-----DDRLVGRRIAFLAT-DGVEKVELEQPRAAILDAGGRVDLL
Mvan_5796|M.vanbaalenii_PYR-1 MPNE---------LQGRKIAFLAA-DGVEKVELEQPRAAVLDAGGDVDLL
MAB_3901|M.abscessus_ATCC_1997 MSDS---------LEGQVVAILAA-DGVERVELEKPREAIPAAGGRVEVL
MSMEG_0536|M.smegmatis_MC2_155 MTHA---------LDGKKVAILAT-DGVERRELVEPREALEQAGARTELL
TH_4363|M.thermoresistible__bu MTTA---------LNGKKVAFLVAPEGVEQVELTEPWKAVEQAGGTPQLV
MLBr_02219|M.leprae_Br4923 MNARIGVITFPGTLDDVDAARAARHVGAEAVSLWHADADLKGVDAVVVPG
: . * . *.* .* .. : ..
MAV_0426|M.avium_104 SLQDGE--IQARNHDLEPAGTFTVDRKVADASVD--------DFDGLVLP
MMAR_5188|M.marinum_M SLQEGD--IQARDHDLEPAGRFAVDRPVQAASVA--------EFDGLVLP
Mflv_1035|M.gilvum_PYR-GCK SVNDGE--IAARNHDLEPAGTFTVDRVVAEATVD--------EYDGLVLP
Mvan_5796|M.vanbaalenii_PYR-1 SVQTGE--IDARNHDLEPAGTFTVDRAVEDARVE--------EYDALVLP
MAB_3901|M.abscessus_ATCC_1997 SPRPGE--IQARDHDLVPAGTVRVDRAVGTASVD--------DYAGLILP
MSMEG_0536|M.smegmatis_MC2_155 SLQTGS--IDARDHDLEPAGTYTVDRLVSEAKGD--------EFDALVVP
TH_4363|M.thermoresistible__bu STEVGK--VQAFNH-LTPADTFDAEVAADSVSAA--------DYAGLVLP
MLBr_02219|M.leprae_Br4923 GFSYGDYLRAGAIARLSPIMTEVVDAVQRGMPVLGICNGFQVLCEAGLLP
. *. . * * .: . ::*
MAV_0426|M.avium_104 GGTVNPD---------KLRLDDTAVSFVRDFVGSGKPVAAICHGPWTLVE
MMAR_5188|M.marinum_M GGTVNPD---------KLRMDKAAVDFVRDFVESGKPVAAICHGPVMLVE
Mflv_1035|M.gilvum_PYR-GCK GGTVNAD---------KLRLEEAAVAFVGEFVRSGKPVAAICHGPWALVE
Mvan_5796|M.vanbaalenii_PYR-1 GGTVNGD---------KLRLSEAAVAFVGDFVRSGKPVAAICHGPWALVE
MAB_3901|M.abscessus_ATCC_1997 GGTVNPD---------KLRIDPGAVRFVHEFVVSGKPVAAICHGPWTLVE
MSMEG_0536|M.smegmatis_MC2_155 GGTVNAD---------KLRSDAKAVAFVHDFVNSGRPVAVICHGPWTLIE
TH_4363|M.thermoresistible__bu GGVANPD---------ILRMHPPAVAFVKSFFDAGKPVASICHAAWTLVE
MLBr_02219|M.leprae_Br4923 GALIRNVGLHFICRDVWLRVISTSTAWTSRFEPETDLLVSLKSGEGRYVA
*. . ** :. :. * :. : . :
MAV_0426|M.avium_104 AG------VAAGRTLTSYPS-IRTDLRNAGAHVVDEEVVVDGN-------
MMAR_5188|M.marinum_M AG------VVRGRTITSYPS-IRTDLRNAGANVVDEEVVVDGN-------
Mflv_1035|M.gilvum_PYR-GCK AE------VASGRTLTSYPS-LRTDLRNAGATVVDQEVCIDGN-------
Mvan_5796|M.vanbaalenii_PYR-1 AD------VVKDRTLTSYPS-LRTDLRNAGATVVDQQVCIDGN-------
MAB_3901|M.abscessus_ATCC_1997 AR------VLAGRTVTSFPS-IRTDLRNAGASVLDEEVVIDHN-------
MSMEG_0536|M.smegmatis_MC2_155 AG------VAKGRTLTSYAS-LRTDLRNAGATVLDEEVVVDGN-------
TH_4363|M.thermoresistible__bu AD------VVRGRTLTSWPS-VKTDLINAGANWVDKEVVVCTDGP----N
MLBr_02219|M.leprae_Br4923 SENVLDELDGEGRVVFRYHDNINGSLRDIAGISSANGRVVGMMPHPEHAI
: .*.: : . :. .* : .. : :
MAV_0426|M.avium_104 -LITSRSPKDLPAFCSTILAQFAGATTG----
MMAR_5188|M.marinum_M -LMTSRSPADLPVFCAAIVKQFAAGACR----
Mflv_1035|M.gilvum_PYR-GCK -LITSRSPRDLDAFCEAITERF----------
Mvan_5796|M.vanbaalenii_PYR-1 -LITSRSPKDLDAFCQAITDQFAGIPART---
MAB_3901|M.abscessus_ATCC_1997 -LITSRSPDDLPAFCQALVDNLAAARIQR---
MSMEG_0536|M.smegmatis_MC2_155 -LITSRSPEDIPAFNKALIDALAQQPQAQAQQ
TH_4363|M.thermoresistible__bu TLVSSRRPDDLPAFCTATLEALAASG------
MLBr_02219|M.leprae_Br4923 EVLTGPSDDGLGLFYSALDSVLAS--------
:::. .: * : :