For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PNETDDRLVGRRIAFLATDGVEKVELEQPRAAILDAGGRVDLLSVNDGEIAARNHDLEPAGTFTVDRVVA EATVDEYDGLVLPGGTVNADKLRLEEAAVAFVGEFVRSGKPVAAICHGPWALVEAEVASGRTLTSYPSLR TDLRNAGATVVDQEVCIDGNLITSRSPRDLDAFCEAITERF*
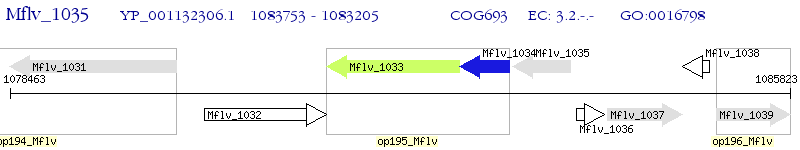
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1035 | - | - | 100% (182) | PfpI family intracellular peptidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3901 | - | 2e-64 | 68.39% (174) | peptidase C56 PfpI |
| M. marinum M | MMAR_5188 | - | 2e-66 | 69.32% (176) | hypothetical protein MMAR_5188 |
| M. avium 104 | MAV_0426 | - | 6e-72 | 74.14% (174) | intracellular protease, PfpI family protein |
| M. smegmatis MC2 155 | MSMEG_0536 | - | 1e-63 | 67.44% (172) | intracellular protease, PfpI family protein |
| M. thermoresistible (build 8) | TH_4363 | - | 2e-42 | 52.54% (177) | PUTATIVE intracellular protease, PfpI family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5796 | - | 2e-83 | 83.52% (182) | PfpI family intracellular peptidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1035|M.gilvum_PYR-GCK MPNETDDRLVGRRIAFLAT-DGVEKVELEQPRAAILDAGGRVDLLSVNDG
Mvan_5796|M.vanbaalenii_PYR-1 MPNE----LQGRKIAFLAA-DGVEKVELEQPRAAVLDAGGDVDLLSVQTG
MMAR_5188|M.marinum_M MSDE----LNGKSIAMLAA-DGVEKVELEQPRSALLQEGARVEVLSLQEG
MAV_0426|M.avium_104 --------MQGKKIAILAA-DGVEKVELEQPAAALREAGAGVEVVSLQDG
MAB_3901|M.abscessus_ATCC_1997 MSDS----LEGQVVAILAA-DGVERVELEKPREAIPAAGGRVEVLSPRPG
MSMEG_0536|M.smegmatis_MC2_155 MTHA----LDGKKVAILAT-DGVERRELVEPREALEQAGARTELLSLQTG
TH_4363|M.thermoresistible__bu MTTA----LNGKKVAFLVAPEGVEQVELTEPWKAVEQAGGTPQLVSTEVG
: *: :*:*.: :***: ** :* *: *. :::* . *
Mflv_1035|M.gilvum_PYR-GCK EIAARNHDLEPAGTFTVDRVVAEATVDEYDGLVLPGGTVNADKLRLEEAA
Mvan_5796|M.vanbaalenii_PYR-1 EIDARNHDLEPAGTFTVDRAVEDARVEEYDALVLPGGTVNGDKLRLSEAA
MMAR_5188|M.marinum_M DIQARDHDLEPAGRFAVDRPVQAASVAEFDGLVLPGGTVNPDKLRMDKAA
MAV_0426|M.avium_104 EIQARNHDLEPAGTFTVDRKVADASVDDFDGLVLPGGTVNPDKLRLDDTA
MAB_3901|M.abscessus_ATCC_1997 EIQARDHDLVPAGTVRVDRAVGTASVDDYAGLILPGGTVNPDKLRIDPGA
MSMEG_0536|M.smegmatis_MC2_155 SIDARDHDLEPAGTYTVDRLVSEAKGDEFDALVVPGGTVNADKLRSDAKA
TH_4363|M.thermoresistible__bu KVQAFNH-LTPADTFDAEVAADSVSAADYAGLVLPGGVANPDILRMHPPA
.: * :* * **. .: . . :: .*::***..* * ** *
Mflv_1035|M.gilvum_PYR-GCK VAFVGEFVRSGKPVAAICHGPWALVEAEVASGRTLTSYPSLRTDLRNAGA
Mvan_5796|M.vanbaalenii_PYR-1 VAFVGDFVRSGKPVAAICHGPWALVEADVVKDRTLTSYPSLRTDLRNAGA
MMAR_5188|M.marinum_M VDFVRDFVESGKPVAAICHGPVMLVEAGVVRGRTITSYPSIRTDLRNAGA
MAV_0426|M.avium_104 VSFVRDFVGSGKPVAAICHGPWTLVEAGVAAGRTLTSYPSIRTDLRNAGA
MAB_3901|M.abscessus_ATCC_1997 VRFVHEFVVSGKPVAAICHGPWTLVEARVLAGRTVTSFPSIRTDLRNAGA
MSMEG_0536|M.smegmatis_MC2_155 VAFVHDFVNSGRPVAVICHGPWTLIEAGVAKGRTLTSYASLRTDLRNAGA
TH_4363|M.thermoresistible__bu VAFVKSFFDAGKPVASICHAAWTLVEADVVRGRTLTSWPSVKTDLINAGA
* ** .*. :*:*** ***.. *:** * .**:**:.*::*** ****
Mflv_1035|M.gilvum_PYR-GCK TVVDQEVCIDGN----LITSRSPRDLDAFCEAITERF----------
Mvan_5796|M.vanbaalenii_PYR-1 TVVDQQVCIDGN----LITSRSPKDLDAFCQAITDQFAGIPART---
MMAR_5188|M.marinum_M NVVDEEVVVDGN----LMTSRSPADLPVFCAAIVKQFAAGACR----
MAV_0426|M.avium_104 HVVDEEVVVDGN----LITSRSPKDLPAFCSTILAQFAGATTG----
MAB_3901|M.abscessus_ATCC_1997 SVLDEEVVIDHN----LITSRSPDDLPAFCQALVDNLAAARIQR---
MSMEG_0536|M.smegmatis_MC2_155 TVLDEEVVVDGN----LITSRSPEDIPAFNKALIDALAQQPQAQAQQ
TH_4363|M.thermoresistible__bu NWVDKEVVVCTDGPNTLVSSRRPDDLPAFCTATLEALAASG------
:*::* : : *::** * *: .* : :