For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PGTDTLLADPRLAAAQRVLGRALVKSVITQAQQRARAGEISPEQVAEHALAALPATASSLRPVINATGVV VHTNLGRAPLSRAALDAVVAAGGATDVEFDLATGRRARRGRGALAALARAVPAAGGVHVVNNNAAALLLV ALTLAPGREIVLSRGELVEIGDGFRIPELLESTGARLREVGTTNRTGLRDYTEALGPQTGFVLKVHPSNF HVTGFTSAVGVAALAAELRTRDIPLVVDIGSGLLAPHPVLPDEPDATSTLADGADLVTASGDKLLGGPQA GLLFGRGDLVERLRRHPAARALRVDKLTLAALEATLTGPPPPVAAALDADVGRLRARAQSLAAALPADAD AVALDCVAAVGGGGAPGVELPSAAVSLPERYAAALRAGRPPVVGRLEGGRCLLDLRTVSPEDDELLAAAV RACSS*
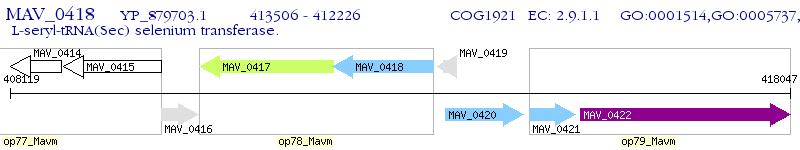
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0418 | selA | - | 100% (426) | selenocysteine synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3726c | - | 8e-05 | 28.88% (277) | hypothetical protein Mb3726c |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv3700c | - | 8e-05 | 28.88% (277) | hypothetical protein Rv3700c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2746c | - | 2e-06 | 28.08% (260) | cysteine desulfurase/aminotransferase |
| M. marinum M | MMAR_5190 | selA | 0.0 | 82.67% (427) | selenocysteine synthase, SelA |
| M. smegmatis MC2 155 | MSMEG_1850 | selA | 0.0 | 80.05% (426) | selenocysteine synthase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2736 | - | 7e-05 | 26.82% (220) | SufS subfamily cysteine desulfurase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0418|M.avium_104 ----------MPGTDTLLADPRLAAAQRVLGRALVKSVITQAQQRARAGE
MMAR_5190|M.marinum_M MGDGSDRRRAVPRTDVLLAHPKLAEAQRTLGRTLVKSVIAQAQQRARAGE
MSMEG_1850|M.smegmatis_MC2_155 ---MTDPRRRVPRTDTLLADPRLAAAEQVLGRALVRSVIAEAQQKARAGE
Mb3726c|M.bovis_AF2122/97 -------------------------MRRSGANSPAGDSLADRWRAARPPV
Rv3700c|M.tuberculosis_H37Rv -------------------------MRRSGANSPAGDSLADRWRAARPPV
MAB_2746c|M.abscessus_ATCC_199 -----------------------MTATVPLDITAIRNDFPILSRTVRGGK
Mvan_2736|M.vanbaalenii_PYR-1 -----------------------MTASVDLDVAAIRADFPILKRVMRGGN
: :. : *
MAV_0418|M.avium_104 ISPEQVAEHALAALPATASSLRPVINATGVVVHTNLGRAPLSRAALDAVV
MMAR_5190|M.marinum_M IEPDQVAEHAIDALPGTASSLRPVINATGVVVHTNLGRAPLSRAALDAVV
MSMEG_1850|M.smegmatis_MC2_155 IPPEQVADAAVAALPTGATSLRPVINATGVVVHTNLGRAPLSRAAIDAVV
Mb3726c|M.bovis_AF2122/97 A---------GLHLDSAACSRQSFAALDAAAQHARH---EAEVGGYVAAE
Rv3700c|M.tuberculosis_H37Rv A---------GLHLDSAACSRQSFAALDAAAQHARH---EAEVGGYVAAE
MAB_2746c|M.abscessus_ATCC_199 P---------LAYLDSGATSQRPVQVLDAERRFLTERNAAVHRGAHQLAE
Mvan_2736|M.vanbaalenii_PYR-1 T---------LAYLDSGATSQKPLQVLDAERQFLTTSYGAVHRGAHQLME
* * * :.. . . ..
MAV_0418|M.avium_104 AAGGATDVEFDLATGRRARRGRGALAALARAVPAAGGVHVVNNNAAALLL
MMAR_5190|M.marinum_M TAGRATDVEFDLATGRRARRGRSALAALARAVPSAAGVHVVNNNAAALLL
MSMEG_1850|M.smegmatis_MC2_155 TASGATDVEFDLETGRRARRGRGALAALARAVPTAGGVHVVNNNAAALLL
Mb3726c|M.bovis_AF2122/97 AAAAVLD------------AGRAAVAALSGLPDAE--VVFTTGSLHALDL
Rv3700c|M.tuberculosis_H37Rv AAAAVLD------------AGRAAVAALSGLPDAE--VVFTTGSLHALDL
MAB_2746c|M.abscessus_ATCC_199 EATDAYE------------GARNAIARFVGVDDGE--IVFTKNATESLNL
Mvan_2736|M.vanbaalenii_PYR-1 ESTDAYE------------QGRADIAAFVGAAPGE--LVFTKNATEALNL
: . : .* :* : : .... :* *
MAV_0418|M.avium_104 VALTLAP-GREIVLSRGELVEIG------DGFRIPELLESTGARLREVGT
MMAR_5190|M.marinum_M TALTLAAPGKEIVLSRGELVEIG------DGFRIPELLASTGSRLREVGT
MSMEG_1850|M.smegmatis_MC2_155 AAMTLAP-GKEMIVSRGELIEIG------DGFRLPALMESTGSRIREVGT
Mb3726c|M.bovis_AF2122/97 LLGSWPGENRTLACLPG------------EYGPNLAVMAAHGFDVRPLPT
Rv3700c|M.tuberculosis_H37Rv LLGSWPGENRTLACLPG------------EYGPNLAVMAAHGFDVRPLPT
MAB_2746c|M.abscessus_ATCC_199 VAYTLGDNRFDRAVGPGDEIVVTELEHHANLVPWQELCRRTGATLRWFGV
Mvan_2736|M.vanbaalenii_PYR-1 VSYVLGDDRSGFAVGPGDVIVVTEMEHHANLIPWQELARRTGATLRWFGL
* : : * :* .
MAV_0418|M.avium_104 TNRTGLRDYTEALGPQTGFVLKVHPSNFHVTGFTSAVGVAALAAELRTRD
MMAR_5190|M.marinum_M TNRTSLRDYAEAIGPNTGFVLKVHPSNFHVTGFTSAVGVS----ELAQLD
MSMEG_1850|M.smegmatis_MC2_155 TNRTHLRDYADAVGPDTGFVLKVHPSNYSVSGFTSSVPVR----ELAALD
Mb3726c|M.bovis_AF2122/97 LQ-DGRVALDDAAFMLADDPPDLVHLTVVASHRGVAQPLAMVAQLCTELK
Rv3700c|M.tuberculosis_H37Rv LQ-DGRVALDDAAFMLADDPPDLVHLTVVASHRGVAQPLAMVAQLCTELK
MAB_2746c|M.abscessus_ATCC_199 TD-DGRIDVDS---LELTEAVKVVAFTHQSNVTGAVAPVAELVRRAKAVG
Mvan_2736|M.vanbaalenii_PYR-1 TD-DGEVDLDS---LKLDERVKVVSFSHHSNVTGVGVPVADMVARAKAVG
: . .: . . :
MAV_0418|M.avium_104 IPLVVDIGSGLLAPHPVLPDEPDATSTLADGADLVTASGDKLLGGPQAGL
MMAR_5190|M.marinum_M APLVVDIGSGLLAPHPLLPDEPDATTMLRDGANLVTASGDKLLGGPQAGL
MSMEG_1850|M.smegmatis_MC2_155 APLVVDIGSGLLTPHPLLPDEPDATTMLRDGADLVTASGDKLLGGPQAGL
Mb3726c|M.bovis_AF2122/97 LPLVVDAAQGLGHVDCAVG----------ADVTYASSR-KWIAGPRGVGV
Rv3700c|M.tuberculosis_H37Rv LPLVVDAAQGLGHVDCAVG----------ADVTYASSR-KWIAGPRGVGV
MAB_2746c|M.abscessus_ATCC_199 ALVVLDACQSVPHMSVNFRE---------LGVDYAAFSGHKMLGPSGVGV
Mvan_2736|M.vanbaalenii_PYR-1 ALTVLDSCQAVPHQPVDFHA---------LDVDFAAFSGHKMLGPTGIGA
*:* ..: . .. .: . : * *
MAV_0418|M.avium_104 LFGRGDLVERLRRHPAARALRVDKLTLA---------------ALEATLT
MMAR_5190|M.marinum_M LFGDADLIERLRRHPAARALRVDKLTLA---------------ALEATVV
MSMEG_1850|M.smegmatis_MC2_155 LFGTEDLIERLRRHPAARALRVDKLTLA---------------ALEATLT
Mb3726c|M.bovis_AF2122/97 LAVRPELMERLRARLPAPDW-MPPLTVA--------QQLGFGEANVAARV
Rv3700c|M.tuberculosis_H37Rv LAVRPELMERLRARLPAPDW-MPPLTVA--------QQLGFGEANVAARV
MAB_2746c|M.abscessus_ATCC_199 LYGRRALLEAMPPFITGGSM-IETVTMEVSTYAPPPQRFEAGVPMTSQVV
Mvan_2736|M.vanbaalenii_PYR-1 LYGRARLLEQLPPFLTGGSM-IETVTMEGATYAPPPQRFEAGTQMISQVV
* *:* : .. : :*: : .
MAV_0418|M.avium_104 GPPPPVAAALDADVGRLRARAQSLAAALPADADAVALDCVAAVGGGGAPG
MMAR_5190|M.marinum_M GPPTPVAQALGADLTELRARAQRITAQLPQSVGATAVDCIAAVGGGGAPG
MSMEG_1850|M.smegmatis_MC2_155 GPPPPVAQALSVDVESLRVRAGRLASLLP---DAKAVDCTAAVGGGGAPD
Mb3726c|M.bovis_AF2122/97 GFSVALGEHLACGPQAIRARLAELGDIARTVLADVSGWRVVEAVDEPSAI
Rv3700c|M.tuberculosis_H37Rv GFSVALGEHLACGPQAIRARLAELGDIARTVLADVSGWRVVEAVDEPSAI
MAB_2746c|M.abscessus_ATCC_199 GLGAAVDYLNAVGMEAVAAHEHQLVSAALAGLAGIEGVRIIGPTDNLDRG
Mvan_2736|M.vanbaalenii_PYR-1 GLGAATRYLGGIGMSAVEEHEHRLVTATLEGLAAIPGVRIIGPAT-AERR
* . . : : :
MAV_0418|M.avium_104 VELPSAAVSLPERYAAALRAGRPPVVGRLEGGRCLLDLR-----------
MMAR_5190|M.marinum_M VELPSAGLSLPESYAATLRTGSPPVVGRLEAGRCLLDLR-----------
MSMEG_1850|M.smegmatis_MC2_155 VELPSAAVSLPEAYAAPLRVGNPSVVGRLENGRCLLDLR-----------
Mb3726c|M.bovis_AF2122/97 TTLAPIDGADPAAVRAWLLSQRRIVTTYAGVERAPLELP---APVLRISP
Rv3700c|M.tuberculosis_H37Rv TTLAPIDGADPAAVRAWLLSQRRIVTTYAGVERAPLELP---APVLRISP
MAB_2746c|M.abscessus_ATCC_199 GAVSFVVDGIHAHDLGQVLDDEGVAVRVGHHCAWPLHRRFGIAASARASF
Mvan_2736|M.vanbaalenii_PYR-1 SPVAFVVDGVHAHDVGQVLDDDGVAVRVGHHCAAPLHRRFGIAATVRASF
:. . . : .. *.
MAV_0418|M.avium_104 --TVSPEDDELLAAAVRACSS------
MMAR_5190|M.marinum_M --TVAPEEDELLLAAVRACLS------
MSMEG_1850|M.smegmatis_MC2_155 --TVAPEDDGKLVEAVRACSS------
Mb3726c|M.bovis_AF2122/97 HVDNTADDLDAFAEALVAATAATSGER
Rv3700c|M.tuberculosis_H37Rv HVDNTADDLDAFAEALVAATAATSGER
MAB_2746c|M.abscessus_ATCC_199 AVYNTLDEVDRLTAGVRRAQEFFS---
Mvan_2736|M.vanbaalenii_PYR-1 AVYNTLDEVDRLIAGVQRAIEFFGRD-
: :: : .: .