For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTIGTRKRVAVVTGASSGIGEATARTLAAQGFHVVAVARRADRITALANQIGGTAIVADVTDDAAVEALA RALSRVDVLVNNAGGAKGLQFVADADLEHWRWMWDTNVLGTLRVTRALLPKLIDSGDGLIVTVTSIAAIE VYDGGAGYTAAKHAQGALHRTLRGELLGKPVRLTEIAPGAVETEFSLVRFDGDQQRADAVYAGMTPLVAA DVAEVIGFVATRPSHVNLDQIVIRPRDQASASRRATHPVR*
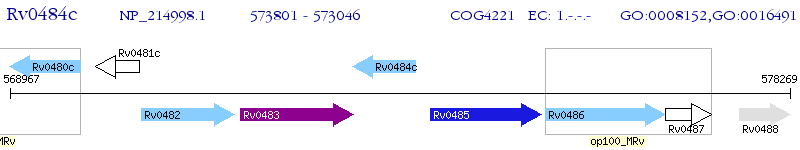
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0484c | - | - | 100% (251) | PROBABLE SHORT-CHAIN TYPE OXIDOREDUCTASE |
| M. tuberculosis H37Rv | Rv1050 | - | 7e-23 | 35.83% (187) | oxidoreductase |
| M. tuberculosis H37Rv | Rv0242c | fabG | 2e-22 | 31.22% (237) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0494c | - | 1e-138 | 99.60% (251) | putative short-chain type oxidoreductase |
| M. gilvum PYR-GCK | Mflv_0088 | - | 1e-102 | 74.49% (243) | short-chain dehydrogenase/reductase SDR |
| M. leprae Br4923 | MLBr_02565 | fabG | 6e-22 | 31.65% (237) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4060 | - | 1e-107 | 79.50% (239) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_0810 | - | 1e-122 | 88.80% (250) | short-chain type oxidoreductase |
| M. avium 104 | MAV_4667 | - | 1e-119 | 88.11% (244) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_0930 | - | 1e-110 | 81.56% (244) | serine 3-dehydrogenase |
| M. thermoresistible (build 8) | TH_0876 | - | 1e-95 | 81.60% (212) | PROBABLE SHORT-CHAIN TYPE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4554 | - | 1e-121 | 88.40% (250) | short-chain type oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_0825 | - | 1e-104 | 78.19% (243) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0088|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0825|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0484c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0494c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MSMEG_0930|M.smegmatis_MC2_155 --------------------------------------------------
TH_0876|M.thermoresistible__bu --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mflv_0088|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0825|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0484c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0494c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MSMEG_0930|M.smegmatis_MC2_155 --------------------------------------------------
TH_0876|M.thermoresistible__bu --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mflv_0088|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0825|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0484c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0494c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MSMEG_0930|M.smegmatis_MC2_155 --------------------------------------------------
TH_0876|M.thermoresistible__bu --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mflv_0088|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0825|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0484c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0494c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MSMEG_0930|M.smegmatis_MC2_155 --------------------------------------------------
TH_0876|M.thermoresistible__bu --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mflv_0088|M.gilvum_PYR-GCK -----MTTPSDARRVAVVTGASAGIGEAIARTLAAQGFHVICVARR--EG
Mvan_0825|M.vanbaalenii_PYR-1 -----MTTPTDARRVAVVTGASAGIGEATARTLAAQGFHVICVARR--EA
Rv0484c|M.tuberculosis_H37Rv -----MTTIGTRKRVAVVTGASSGIGEATARTLAAQGFHVVAVARR--AD
Mb0494c|M.bovis_AF2122/97 -----MTTIGTRKRVAVVTGASSGIGEATARTLAAQGFHVVAVARR--AD
MMAR_0810|M.marinum_M -----MTTTDAGNRVAVVTGASSGIGEATARTLAAQGFHVVAVARR--AD
MUL_4554|M.ulcerans_Agy99 -----MTTTDAGNRVAVVTGASSGIGEATARTLAAQGFHVVAVARR--AD
MAV_4667|M.avium_104 -----MVGTGRNERVAVVTGASSGIGEATARTLAAQGFHVVAVARR--AK
MSMEG_0930|M.smegmatis_MC2_155 -----MSTPHSDRPVAVVTGASAGIGAATAKTLASLGFHVICAARR--AD
TH_0876|M.thermoresistible__bu ------------------------------------------VARR--AE
MAB_4060|M.abscessus_ATCC_1997 -----MTNPTFDGRVAVVTGASSGIGAATAKSLAALGFHVVVAARR--ED
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
.
Mflv_0088|M.gilvum_PYR-GCK PIKALAAEIDGTAIVADVTDPTAVAALAGRLD-----RVDVLVNNAGGAR
Mvan_0825|M.vanbaalenii_PYR-1 PIQALAAEIDGTAIVADVTDPTAVAALAGRLD-----RVDVLVNNAGGAR
Rv0484c|M.tuberculosis_H37Rv RITALANQIGGTAIVADVTDDAAVEALARALS-----RVDVLVNNAGGAK
Mb0494c|M.bovis_AF2122/97 RITALANQIGGTAIVADVTDDAAVEALARALS-----RVDVLVNNAGGAK
MMAR_0810|M.marinum_M RITALTDQIGGTPIVADVTDGAAVAALADGLS-----RVDVLVNNAGGAK
MUL_4554|M.ulcerans_Agy99 RITALTDQIGGTPIVADVTDGAAVAALADGLS-----RVDVLVNNAGGAK
MAV_4667|M.avium_104 RIHALANEIGGTAIVADVTDDAAVAALASKLS-----RVDVLVNNAGGAR
MSMEG_0930|M.smegmatis_MC2_155 RIDALAKEIDGTAVVADVTDDESVNALAAQLD-----RVDVLVNNAGGAK
TH_0876|M.thermoresistible__bu LVEALAAEISGTASVTDVTDRAAVEALADQLD-----RVDVLVNNAGGAK
MAB_4060|M.abscessus_ATCC_1997 RVKSLAGEIGGTAFVLDVTDADSVAALTQSLD-----RVDVLVNNAGGAW
MLBr_02565|M.leprae_Br4923 ALDETANRVGGTTLSLDVTADDAVDKITEHLHDYQGGHADILVNNAG-IT
: : .:.**. *** :* :: * :.*:******
Mflv_0088|M.gilvum_PYR-GCK GLESISDADVENWRWMWEANVMGTLHVTRALLPKLIDSGDGLIVTITSIA
Mvan_0825|M.vanbaalenii_PYR-1 GLESVADADVEHWRWMWEANVLGTLHVTRALLPKLIGSGDGLIVTVTSIA
Rv0484c|M.tuberculosis_H37Rv GLQFVADADLEHWRWMWDTNVLGTLRVTRALLPKLIDSGDGLIVTVTSIA
Mb0494c|M.bovis_AF2122/97 GLQFVADADLEHWRWMWDTNVLGTLRVTRALLPKLIDSGDGLIVTVTSIA
MMAR_0810|M.marinum_M GLEPVADADLEHWRWMWETNVLGTLRVTRALLPKLIASGDGLIVTVTSIA
MUL_4554|M.ulcerans_Agy99 ELEPVADADLEHWRWMWETNVLGTLRVTRALLPKLIASGDGLIVTVTSIA
MAV_4667|M.avium_104 GLEPVAEADLEHWRWMWETNVIGTLRVTRALLPKLVDSGDGLIVTVTSVA
MSMEG_0930|M.smegmatis_MC2_155 GLAPVSDADLDHWRWMWETNVLGTLRVTRALLPKLIDSGDGLIVTVTSIA
TH_0876|M.thermoresistible__bu GLEPVAEADLDHWRWMWETNVLGTLHVTRALLPKLIASGDGLIVTVTSIA
MAB_4060|M.abscessus_ATCC_1997 GLEPVLEADLEHWRWMWETNVVGTLQVTRALLPKLIDSGDGLIVTVTSIA
MLBr_02565|M.leprae_Br4923 RDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGGRVIVLSSMA
: : * .* : :.*::.. ::*..*: : * .* ::.::*:*
Mflv_0088|M.gilvum_PYR-GCK AVEIYDNGGGYTSAKHAQGVLHRTLRSELLGKPVRLTEVAPGMVKTDFSL
Mvan_0825|M.vanbaalenii_PYR-1 AVEIYDNGAGYTSAKHAQGVLHRTLRSELLGKPVRLTEVAPGMVKTEFSL
Rv0484c|M.tuberculosis_H37Rv AIEVYDGGAGYTAAKHAQGALHRTLRGELLGKPVRLTEIAPGAVETEFSL
Mb0494c|M.bovis_AF2122/97 ALEVYDGGAGYTAAKHAQGALHRTLRGELLGKPVRLTEIAPGAVETEFSL
MMAR_0810|M.marinum_M ALEIYDGGAGYTAAKHAQGALHRTLRGELLGLPVRLTEIAPGAVETEFSL
MUL_4554|M.ulcerans_Agy99 ALEIYDGGAGYTAAKHAQGALHRTLRGELLGLPVRLTEIAPGAVETEFSL
MAV_4667|M.avium_104 ALEVYDGGAGYTAAKHGQGALHRTLRGELLGKPVRLTEIAPGAVETEFSL
MSMEG_0930|M.smegmatis_MC2_155 AFETYDGGAGYTSAKHAQGALHRTLRGELLGKPVRLTEIAPGAVETEFSL
TH_0876|M.thermoresistible__bu AFEIYDGGAGYTAAKHAQSALHRTLRGELLGKPVRLTEIQPGMVETEFSL
MAB_4060|M.abscessus_ATCC_1997 ALDVYDGGSGYTSAKHAQAALHRTLRGELLGKPVRLTEVAPGAVETEFSV
MLBr_02565|M.leprae_Br4923 GIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGFIETQMTA
.. . .*::** . * ::* * : :. : ** ::*:::
Mflv_0088|M.gilvum_PYR-GCK NRFDGDAERAEKVYEGVTPLVAEDIAEVVGFVASRPSHVDLDLIVVRPRD
Mvan_0825|M.vanbaalenii_PYR-1 NRFDGDAERADKVYQGVTPLVAEDIAEVIGFVASRPSHVDLDLIVVRPRD
Rv0484c|M.tuberculosis_H37Rv VRFDGDQQRADAVYAGMTPLVAADVAEVIGFVATRPSHVNLDQIVIRPRD
Mb0494c|M.bovis_AF2122/97 VRFDGDQQRADAVYAGMTPLVAADVAEVIGFVATRPSHVNLDQIVIRPRD
MMAR_0810|M.marinum_M VRFDGDEQRADAVYSGMTPLVAADIAEVIGFVASRPPHVNLDQIIIRPRD
MUL_4554|M.ulcerans_Agy99 VRFDGDEQRADAVYSGMTPLVAADIAEVIGFVASRPPHVNLDQIIIRPRD
MAV_4667|M.avium_104 VRFDGDQQRADAVYTGITPLVAADVAEVIGFVASRPAHVNLDLIVMKPRD
MSMEG_0930|M.smegmatis_MC2_155 VRFDGDRERADNVYKGITPLVAEDIAEVIGFVASRPSHVNLDQIVIRPRD
TH_0876|M.thermoresistible__bu VRFDGDREKADAVYQGLTPLVAEDIAEVIGFAASRPPHVNLDQIVIRPRD
MAB_4060|M.abscessus_ATCC_1997 VRFDGDQDRADAVYRGITPLVAEDVAEIIAFVASRPPHVNLDQIVVKPRD
MLBr_02565|M.leprae_Br4923 AIPLATREVGRRLNSLLQGGLPVDVAETIAYFA-IPASNAVTGNVIRVCG
. : . : : :. *:** :.: * *. : ::: .
Mflv_0088|M.gilvum_PYR-GCK QVSGASGSRFHRQS--
Mvan_0825|M.vanbaalenii_PYR-1 QVSGAAGSRFNRST--
Rv0484c|M.tuberculosis_H37Rv QAS--ASRRATHPVR-
Mb0494c|M.bovis_AF2122/97 QAS--ASRRATHPVR-
MMAR_0810|M.marinum_M QAS--ATRRANHQV--
MUL_4554|M.ulcerans_Agy99 QAS--ATRRANHQV--
MAV_4667|M.avium_104 QAS--ASRFNRRG---
MSMEG_0930|M.smegmatis_MC2_155 QAP--NGR-FNRRT--
TH_0876|M.thermoresistible__bu QAS--ATRKFTRPAGE
MAB_4060|M.abscessus_ATCC_1997 QAS--STRRATRQ---
MLBr_02565|M.leprae_Br4923 QAMLGA----------
*.