For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGEASTGVPGTKIALALGAGGARGYAHIGVIQELHERGFEISGISGSSMGALVGGLEAAGALDEFALWA TSLTQRAVLRLLDPTWTSPGFFRAEKILDVVRELLGDVAIEDLPIPFTAVATDLIAGKSVWLQNGSLASA IRASIAIPGMISPHVLNGRVLADGGVLDLPAVAPLAGVHAELRVAVTLSGGDERRAPAPPEPEPEPREPR VTTEWLNRLLRSTSALFSTDDPEVAGQPDPAAKLTSFEVMNRTIEVMQSQLARMQLAAHPPDVLIEVPRS VSRSLDFHRATEVIEVGRRCAAEALDAYAHTN
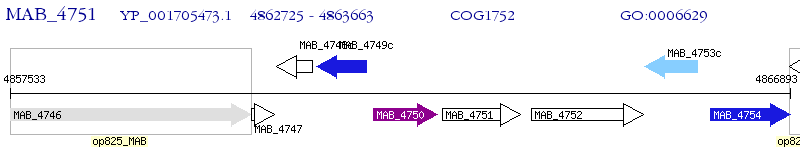
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4751 | - | - | 100% (312) | hypothetical protein MAB_4751 |
| M. abscessus ATCC 19977 | MAB_4758 | - | 2e-14 | 34.50% (171) | patatin-like protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1092c | - | 9e-95 | 54.94% (344) | hypothetical protein Mb1092c |
| M. gilvum PYR-GCK | Mflv_2030 | - | 5e-96 | 57.27% (330) | patatin |
| M. tuberculosis H37Rv | Rv1063c | - | 9e-95 | 54.94% (344) | hypothetical protein Rv1063c |
| M. leprae Br4923 | MLBr_01423 | - | 4e-05 | 24.49% (196) | hypothetical protein MLBr_01423 |
| M. marinum M | MMAR_4403 | - | 2e-94 | 55.17% (348) | hypothetical protein MMAR_4403 |
| M. avium 104 | MAV_1190 | - | 8e-93 | 55.29% (340) | patatin |
| M. smegmatis MC2 155 | MSMEG_5284 | - | 2e-96 | 57.78% (334) | patatin |
| M. thermoresistible (build 8) | TH_0679 | - | 9e-95 | 58.81% (318) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0218 | - | 5e-94 | 54.89% (348) | hypothetical protein MUL_0218 |
| M. vanbaalenii PYR-1 | Mvan_4687 | - | 3e-96 | 57.98% (326) | patatin |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5284|M.smegmatis_MC2_155 --------MTHNESMRVALALGSGSARGYAHIGVINELRARGYEIVGISG
TH_0679|M.thermoresistible__bu ----------------VALALGSGGARGYAHIGVIEELRARGHEVVGVSG
Mflv_2030|M.gilvum_PYR-GCK --------------MRVALALGSGGARGYAHIGVLNELRERGHEVVGIAG
Mvan_4687|M.vanbaalenii_PYR-1 --------------MRVALALGSGGARGYAHIGVLNELTERGHEVVGIAG
Mb1092c|M.bovis_AF2122/97 MPAPAALRVRGSSSPRVALALGSGGARGYAHIGVIQALRERGYDIVGIAG
Rv1063c|M.tuberculosis_H37Rv MPAPAALRVRGSSSPRVALALGSGGARGYAHIGVIQALRERGYDIVGIAG
MMAR_4403|M.marinum_M MTGPIAVRTHGSSSTRVALALGSGGARGYAHIGVIEALRERGYEIAGIAG
MUL_0218|M.ulcerans_Agy99 MTGPIAVRTHGSSSTRVALALGSGGARGYAHIGVIEALRERGYEIAGIAG
MAV_1190|M.avium_104 ----------------MALALGSGGARGYAHIGVIEALQARGYDIVGVAG
MAB_4751|M.abscessus_ATCC_1997 ---MTGEASTGVPGTKIALALGAGGARGYAHIGVIQELHERGFEISGISG
MLBr_01423|M.leprae_Br4923 -----MLYLVIVSVVHADLVCQGGGIRGIGLVGAVDALATAGYRFPRVAG
*. .*. ** . :*.:: * *. . ::*
MSMEG_5284|M.smegmatis_MC2_155 SSMGALVGGLYAAGKLD--EFAEWACTLTQRAVLRLLDPSLSAAGILRAE
TH_0679|M.thermoresistible__bu SSMGALVGGVMAAGKLD--DYAAWARTLTQRSVLRLLDPSLTAPGVLRAE
Mflv_2030|M.gilvum_PYR-GCK SSMGALVGGLEAAGKTD--EFTLWATSLTQRAVLRLLDPSISSPGVLRAE
Mvan_4687|M.vanbaalenii_PYR-1 SSMGALVGGLQAAGKLD--EYTAWASSLTQRAVLRLLDPSITSAGVLRAE
Mb1092c|M.bovis_AF2122/97 SSMGAVVGGVHAAGRLD--EFAHWAKSLTQRTILRLLDPSISAAGILRAE
Rv1063c|M.tuberculosis_H37Rv SSMGAVVGGVHAAGRLD--EFAHWAKSLTQRTILRLLDPSISAAGILRAE
MMAR_4403|M.marinum_M SSMGAVVGGLQAAGRLG--EFSDWAKSLNQRTILRLLDPSITAAGVLRAE
MUL_0218|M.ulcerans_Agy99 SSMGAVVGGLQAAGRLG--EFSDWAKSLNQRTILRLLDPSITAAGVLRAE
MAV_1190|M.avium_104 SSMGALVGGLQAAGRLD--EFAEWAKSLTQRTILRLLDPSISAAGVMRAG
MAB_4751|M.abscessus_ATCC_1997 SSMGALVGGLEAAGALD--EFALWATSLTQRAVLRLLDPTWTSPGFFRAE
MLBr_01423|M.leprae_Br4923 TSAGALVASLIAALQTAGEPLTRLAEVMRTIDYRKFFDRSLIG----RVP
:* **:*..: ** : * : :::* : . *.
MSMEG_5284|M.smegmatis_MC2_155 KILDAVRDILGDVTIEELPIPYTAVATDLITGKAVWLQRGRVDTAIRASI
TH_0679|M.thermoresistible__bu KILDAVREILGEATIETLPIPFTAVATDLIAGKSVWLQRGPVDGAIRASI
Mflv_2030|M.gilvum_PYR-GCK KILESVRDILGEVTIEDLPIPYTSVATDLITGKSVWMQRGRVDDAIRASI
Mvan_4687|M.vanbaalenii_PYR-1 KILDAVRAIVGDVTIEELPIPYTAVATDLITGKSVWMQRGPLDDAIRASI
Mb1092c|M.bovis_AF2122/97 KILDAVRDIVGPVAIEQLPIPYTAVATDLLAGKSVWFQRGPLDAAIRASI
Rv1063c|M.tuberculosis_H37Rv KILDAVRDIVGPVAIEQLPIPYTAVATDLLAGKSVWFQRGPLDAAIRASI
MMAR_4403|M.marinum_M KILDAVRDILGPVTIEQLPVPYTAVATDILAGKSVWFQRGPLDEAIRASI
MUL_0218|M.ulcerans_Agy99 KILDAVRDILGPVTIEQLPVPYTAVATDILAGKSVWFQRGPLDEAIHASI
MAV_1190|M.avium_104 KILDAVRDILGPVAIEDLRIPYTAVATDLLAGKSVWFQRGPLDEAIRASI
MAB_4751|M.abscessus_ATCC_1997 KILDVVRELLGDVAIEDLPIPFTAVATDLIAGKSVWLQNGSLASAIRASI
MLBr_01423|M.leprae_Br4923 LIGGGLSLLVSDGVYQGAYL--EEWLTSLLGDLGVYTFGDLRTGEEPEQF
* : ::. . : : *.:: . .*: . .:
MSMEG_5284|M.smegmatis_MC2_155 AIPGVIAPHVHDGRLLGDGGILDPLPMAPIASVNADLTIAVSVSGSETPP
TH_0679|M.thermoresistible__bu AIPGVIAPHVLDGRLLADGGILDPVPMAPIAAVNADVTIAVSLAGSEAG-
Mflv_2030|M.gilvum_PYR-GCK AIPGLITPHVLGGRLLADGGILDPLPMAPIAAAPADITIAVSLSGEDPA-
Mvan_4687|M.vanbaalenii_PYR-1 AIPGVITPYVLDGRLLADGGILDPLPMAPIAAVAADMTIAVSISGDDPE-
Mb1092c|M.bovis_AF2122/97 AIPGVIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSEAG-
Rv1063c|M.tuberculosis_H37Rv AIPGVIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSEAG-
MMAR_4403|M.marinum_M AIPGIIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSESG-
MUL_0218|M.ulcerans_Agy99 AIPGIIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSESG-
MAV_1190|M.avium_104 AIPGVIAPHTLDGRLLADGGILDPLPMAPLSAVNADLTIAVNLSGSEAI-
MAB_4751|M.abscessus_ATCC_1997 AIPGMISPHVLNGRVLADGGVLDLPAVAPLAGVHAELRVAVTLSGGDER-
MLBr_01423|M.leprae_Br4923 AWSLVVTASDLSRRRLVR----IPWDLDAYGINPDDFSVARAVHASSAIP
* . :::. . * * : . . :. :* : . .
MSMEG_5284|M.smegmatis_MC2_155 ETASEAHSEPAE-GGGTKPTTEWLNRMMRSTSAVLDTAAVRSVLDRPTSR
TH_0679|M.thermoresistible__bu ------GAEPADRGAGARPTAEWVN---RLTAALLDSAAARTVRDR----
Mflv_2030|M.gilvum_PYR-GCK --------VDRRMQDLRQPTTDWLSRMWRSSSSLLDTKAARSLLETPTGR
Mvan_4687|M.vanbaalenii_PYR-1 --------ASGRGRESRAPSTDWLNRMWRSSSSLLDTKAARSLLDTPTGR
Mb1092c|M.bovis_AF2122/97 ----------PARDAEPNVTAEWLNRMVRSTSALFDVSAARSLLDRPTAR
Rv1063c|M.tuberculosis_H37Rv ----------PARDAEPNVTAEWLNRMVRSTSALFDVSAARSLLDRPTAR
MMAR_4403|M.marinum_M ----------PSRESEPGVTTEWLNRMVRTTSSIFDATAARSLLDRPT-R
MUL_0218|M.ulcerans_Agy99 ----------PSRESEPGVTTEWLNRMVRTTSSIFDATAARSLLDRPT-R
MAV_1190|M.avium_104 ----------TRREAEPAATAEWLTRMMRSTSALFDTAAARSLLDRPTAR
MAB_4751|M.abscessus_ATCC_1997 --RAPAPPEPEPEPREPRVTTEWLNRLLRSTSALFSTDDP----------
MLBr_01423|M.leprae_Br4923 ----------YVFEPVQVAGATWVDGGLLSTFPVELFDRSDGDARWPTFG
: *: : .:
MSMEG_5284|M.smegmatis_MC2_155 AVLSRFGVSVTESD-DGTDASEDA----TVPESSAEP--AG----VPRLG
TH_0679|M.thermoresistible__bu -----FGSGAAPADPDPVDQDREL----EEAAENSRS--IT----VPRLG
Mflv_2030|M.gilvum_PYR-GCK ---AVLGRFVAADD-EPIDAPPEV----VKHDSADATGVPE----LPRLG
Mvan_4687|M.vanbaalenii_PYR-1 ---AVLGRFAAGDD-EPLPEPDDK----VTEEPGNAG--------IPKLG
Mb1092c|M.bovis_AF2122/97 AVLSRFGAAAAESDSWSQAPEIEQRPAGPPADREEAADTPG----LPKMG
Rv1063c|M.tuberculosis_H37Rv AVLSRFGAAAAESDSWSQAPEIEQRPAGPPADREEAADTPG----LPKMG
MMAR_4403|M.marinum_M AVLNRFGAGSPDQDSWPEDAQIEA----PVPERDDGGYTPG----LPKMG
MUL_0218|M.ulcerans_Agy99 AVLNRFGAGSPDQDSWPEDAQIEA----PVPERDDGGYTPG----LPKMG
MAV_1190|M.avium_104 AVLSRLGGPSGDADAWPDNPEEPDQP-VLADDDDEAVELPGAATDVPKLG
MAB_4751|M.abscessus_ATCC_1997 -------EVAGQPD-----P-------------------------AAKLT
MLBr_01423|M.leprae_Br4923 ---IRLSARPGIPPTHPVRGPVSLG---IAAIETLVSNQDN-----AYID
. :
MSMEG_5284|M.smegmatis_MC2_155 SFEVMYRTIDIAQAALARHTLAAYPPDLLIEVPRTVCRSLEFNRAAEVIE
TH_0679|M.thermoresistible__bu SFEVMNRTIDIAQAALARHTLAAYPPDVVIEIPRSVCRSLDFHRAAEVID
Mflv_2030|M.gilvum_PYR-GCK SFEVMNRVIDIAQSALARHTLAAYPPDLLIEVPRTVCRSLDFHRAHEVIE
Mvan_4687|M.vanbaalenii_PYR-1 AFEVMNRVIDIAQAALARHTLAAYPPDLLIEVPRTVCRSLDFHRATEVIE
Mb1092c|M.bovis_AF2122/97 SFEVMNRTIDIAQSALARHTLAGYPADLLIEVPRSTCRSLEFHRAVEVIA
Rv1063c|M.tuberculosis_H37Rv SFEVMNRTIDIAQSALARHTLAGYPADLLIEVPRSTCRSLEFHRAVEVIA
MMAR_4403|M.marinum_M SFEVMNRTIDIAQSALARHTLAAYPADLLIEVPRSTCRSLEFHRASEVIA
MUL_0218|M.ulcerans_Agy99 SFEVMNRTIDIAQSALARHTLAAYPADLLIEVPRSTCRSLEFHRASEVIA
MAV_1190|M.avium_104 SFEVMNRTIDIAQAALARHTLAVYPPDLLIEVPRSICRGLEFHRAVEVID
MAB_4751|M.abscessus_ATCC_1997 SFEVMNRTIEVMQSQLARMQLAAHPPDVLIEVPRSVSRSLDFHRATEVIE
MLBr_01423|M.leprae_Br4923 DSCTMLRTIFVPADEVSPIDFNITVEQRQALYQSGLQAGQQFLKTWNYTE
.* *.* : :: : : . :* :: :
MSMEG_5284|M.smegmatis_MC2_155 TGRRLAAEALDS---------------
TH_0679|M.thermoresistible__bu AGRELAAAALDDAGLT-----------
Mflv_2030|M.gilvum_PYR-GCK IGQELASAALEALDPEPSEA-------
Mvan_4687|M.vanbaalenii_PYR-1 IGQELASAALDTLEQKVPETASDT---
Mb1092c|M.bovis_AF2122/97 VGRALATQALEAFEIDDDESAAATIEG
Rv1063c|M.tuberculosis_H37Rv VGRALATQALEAFEIDDDESAAATIEG
MMAR_4403|M.marinum_M VGRALANRALDALES-ADQPATPAIEG
MUL_0218|M.ulcerans_Agy99 VGRALANRALDALES-ADQPATPAIEG
MAV_1190|M.avium_104 AGRALADQALDALEDGDDDR--PAVER
MAB_4751|M.abscessus_ATCC_1997 VGRRCAAEALDAYAHTN----------
MLBr_01423|M.leprae_Br4923 YLTACGRPVTRSAGL------------
. .