For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMERLREWFRAMHVLGPRWSVFVASMVGINAATLLLLFYFLRHWMPFKRPAGDWVFENGPPLFWLLFGIM IVTSVGWALHMEHRLARWYCRGGPPTALELRTALTAPLQQSLLHLGGWAIGATVFIAAGAMHDSKWTLAT IIGCVQSATASFGITYLLGERILRPIAAKALSASEFHGRFAPSVFVRVRLSWWIGTLAPAIGIIALCGMA LWKPEDLTSTRDLALTVLLIVSTTLVGSYGLALLTAGQLADPVRQLRTAISDVAQGDFNARVEVYDGSEL GVLQAGFNRMMQVGEERRQLRELFGKHVGADVASQALQLGTNLGGENRYVAVFFVDMIGSTSMAADRDPE EVLTILNDFFRIVVEVVDSRGGFVNKFVGDEALSVFGAPLHRPDATTACLAAARELRDRLNEDFPHPFEF AIGVSAGLAVAGNVGATARYEYSVIGDPVNEASRLTELAKHRPSRLLASTNALYFADPDEQKHWEHGDSV LLRGRTRATHLAWPAGT
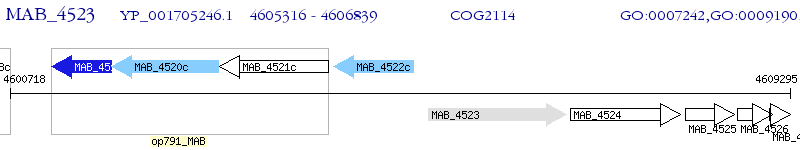
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4523 | - | - | 100% (507) | putative adenylate cyclase |
| M. abscessus ATCC 19977 | MAB_0490c | - | 7e-95 | 41.30% (506) | putative adenylate cyclase |
| M. abscessus ATCC 19977 | MAB_1459c | - | 4e-86 | 39.51% (486) | hypothetical protein MAB_1459c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3669 | - | 3e-95 | 42.97% (498) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1395 | - | 6e-95 | 42.77% (498) | putative adenylate/guanylate cyclase |
| M. tuberculosis H37Rv | Rv3645 | - | 3e-95 | 42.97% (498) | transmembrane protein |
| M. leprae Br4923 | MLBr_00201 | - | 2e-91 | 42.49% (506) | hypothetical protein MLBr_00201 |
| M. marinum M | MMAR_5137 | - | 2e-95 | 42.19% (512) | adenylate cyclase |
| M. avium 104 | MAV_0518 | - | 3e-93 | 42.30% (513) | adenylate cyclase, family protein 3 |
| M. smegmatis MC2 155 | MSMEG_6154 | - | 5e-93 | 42.48% (499) | adenylate cyclase, family protein 3 |
| M. thermoresistible (build 8) | TH_0858 | - | 4e-93 | 44.31% (501) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4221 | - | 3e-95 | 42.19% (512) | adenylate cyclase |
| M. vanbaalenii PYR-1 | Mvan_5395 | - | 4e-96 | 43.20% (500) | putative adenylate/guanylate cyclase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3669|M.bovis_AF2122/97 MDAEAFVGFRQVPAARYGGLMATTAA-LPRRIHAFVR--WVVRTPWPLFS
Rv3645|M.tuberculosis_H37Rv MDAEAFVGFRQVPAARYGGLMATTAA-LPRRIHAFVR--WVVRTPWPLFS
MLBr_00201|M.leprae_Br4923 --------------------MATQAAGLPGRIGAFVR--WGIRTPWPLFS
MMAR_5137|M.marinum_M --------------------MATVAT-LPGRINGFAR--WVVRTPWPVFS
MUL_4221|M.ulcerans_Agy99 --------------------MATVAT-LPGRINGFAR--WVVRTPWPVFS
MAV_0518|M.avium_104 --------------------MTTAAA-LPGRISAFAR--WVVRTPWPLFW
Mflv_1395|M.gilvum_PYR-GCK --------------------MAADPT-ETGRISAFVR--WVARTPWPVFT
Mvan_5395|M.vanbaalenii_PYR-1 --------------------MAAEPI-EVGRISAFVR--WVARTPWPVFT
MSMEG_6154|M.smegmatis_MC2_155 --------------------MTTEPI-PIARIGAFAR--WVARTPWPVFT
TH_0858|M.thermoresistible__bu -----------------------------------------------VFA
MAB_4523|M.abscessus_ATCC_1997 ---------------------------MMERLREWFRAMHVLGPRWSVFV
:*
Mb3669|M.bovis_AF2122/97 LSMLQSDIIGALFVLGFLRYGLP---PQDNIQLQDLPPVNLLIFVSTVII
Rv3645|M.tuberculosis_H37Rv LSMLQSDIIGALFVLGFLRYGLP---PQDNIQLQDLPPVNLLIFVSTVII
MLBr_00201|M.leprae_Br4923 LSMLQSDIIGALFVLGFLRYGLP---PQDRVELQDLPIRNLAISTLSVVV
MMAR_5137|M.marinum_M LSMLQADIIGGLFVLGFLRYGLP---PNDRIQLQDLPPLNLAIFLTSVIT
MUL_4221|M.ulcerans_Agy99 LSMLQADIIGGLFVLGFLRYGLP---PNDRIQLQDLPPLNLAIFLTSVIT
MAV_0518|M.avium_104 LSMMQADIIGALFVLGFLRYALP---PEDRIQLQDLPVVNLAVFATSLVV
Mflv_1395|M.gilvum_PYR-GCK LGMLQADIIGALLVLGFLRFGLP---PEDRIQLQDLPAYNLAIFLGYLFV
Mvan_5395|M.vanbaalenii_PYR-1 LGMLQADIIGALLVLGFLRFGLP---PEDRIQLQDLPAYNLAIFLGYLFI
MSMEG_6154|M.smegmatis_MC2_155 LGMLQADIIGALFVLGFLRFGLP---PEDRIQLQDLPAFNLAIFLAYLFV
TH_0858|M.thermoresistible__bu LGMVQADIIGALFVLGFLRFGLP---PGDRLELQDLPAFNLALFLGYLVV
MAB_4523|M.abscessus_ATCC_1997 ASMVGINAATLLLLFYFLRHWMPFKRPAGDWVFENGPPLFWLLFG----I
.*: : *::: ***. :* * . ::: * :
Mb3669|M.bovis_AF2122/97 LFLAGAVVNLKLLMPVFRWQRRDNLLTEPDPAATELARSRALRMPLYRTL
Rv3645|M.tuberculosis_H37Rv LFLAGAVVNLKLLMPVFRWQRRDNLLTEPDPAATELARSRALRMPLYRTL
MLBr_00201|M.leprae_Br4923 AFSTGFAVNLKLLMPVFRWQRRDNRLAEADPAATELARSRALRMPFYHTI
MMAR_5137|M.marinum_M LFLAGFALNLKLLVPVFRWQRRDNLVAAADPSATELARSRVLRMPFYRTL
MUL_4221|M.ulcerans_Agy99 LFLAGFALNLKLLVPVFRWQRRDNLVAAADPSATELARSRVLRMPFYRTL
MAV_0518|M.avium_104 LFLAASVVNLTLLMPVFRWQRRDNLLAESDPAATELARIRALRMPFYRTV
Mflv_1395|M.gilvum_PYR-GCK SFTVASYLTLRMLIPVMRWQRRDMLLGDRDPSDTEVARMRALKMPFYRSL
Mvan_5395|M.vanbaalenii_PYR-1 SFTVASYVTLRMLIPVMRWQRREMLLGDRDPAETEVARMRALKMPFYRSL
MSMEG_6154|M.smegmatis_MC2_155 SFTVGAYLALRLLIPVIRWQRRETVLSEGDPGITELARARALKMPFYRTL
TH_0858|M.thermoresistible__bu SFTVGAYVTLRLLLPVIRWQRRDTLLADPDPAATELARARALRMPFYRTL
MAB_4523|M.abscessus_ATCC_1997 MIVTSVGWALHMEHRLARWYCRG-----GPPTALELR--TALTAPLQQSL
: .. * : : ** * * *: .* *: :::
Mb3669|M.bovis_AF2122/97 ISLAVWATGGGVFILASWSVAKHAAPVVAVATALGATATAIIGYLQSERV
Rv3645|M.tuberculosis_H37Rv ISLAVWATGGGVFILASWSVAKHAAPVVAVATALGATATAIIGYLQSERV
MLBr_00201|M.leprae_Br4923 ISLVTWGIGGAVFIIASWSAARHSAPIVALATALGATATAIIGYLQSERV
MMAR_5137|M.marinum_M ISVGSWAIGSVIFIVASWSVARYAAPVVAIATALGAIATAIIGYLQSERV
MUL_4221|M.ulcerans_Agy99 ISVGSWAIGSVIFIVASWSVARYAAPVVAIATALGAIATAIIGYLQSERV
MAV_0518|M.avium_104 TSMVIWCIGGVVFIIASWSVARFAAPVVAVATGLGAAATAIIGYLQCERV
Mflv_1395|M.gilvum_PYR-GCK ISATNWLLGSVVFIVASWPVASKSAPVVAVATGLGATATAIIGYLQSERV
Mvan_5395|M.vanbaalenii_PYR-1 ISATNWLLGSVVFIVASWPVASKSAPVVAVATALGATATAIIGYLQSERV
MSMEG_6154|M.smegmatis_MC2_155 ISVTNWCLGSIVFIVASWPVASHSAPVVAVATALGATATAIIGYLQSERV
TH_0858|M.thermoresistible__bu ISGTNWFLGTIVFIVASWPVAGHSAPVIAVATGLGATATAIIGYLQAERV
MAB_4523|M.abscessus_ATCC_1997 LHLGGWAIGATVFIAAGAMHDSKWTLATIIGCVQSATASFGITYLLGERI
* * :** *. : :. .* *: * ** **:
Mb3669|M.bovis_AF2122/97 LRPVAVAALRSGVPENVNAPGVILRLMLAWIPSTGVPLLAIVLAVAADKI
Rv3645|M.tuberculosis_H37Rv LRPVAVAALRSGVPENVNAPGVILRLMLAWIPSTGVPLLAIVLAVAADKI
MLBr_00201|M.leprae_Br4923 LRPVAVAALRSGVPENVKTPGVTLRLMLTWVPSTGVPVLAIVLAVVADKI
MMAR_5137|M.marinum_M LRPVAVAALRSGVPENLRSPGVILRLMLAWVLSTAVPLLAIVLAVVADKM
MUL_4221|M.ulcerans_Agy99 LRPVAVAALRSGVPENLRSPGVISRLMLAWVLSTAVPLLAIVLAVVADKM
MAV_0518|M.avium_104 LRPVAVAALRSGVPENVKAPGVILRQILTWMLSTAVPVLAIVLAVVADKT
Mflv_1395|M.gilvum_PYR-GCK LRPVAVAALRGGVPENFRAPGVILRQVLTWVLSTGVPVLAIVLALVASKF
Mvan_5395|M.vanbaalenii_PYR-1 LRPVAVAALRGGVPENFRAPGVILRQVLSWVLSTGVPVLAILLALIASKL
MSMEG_6154|M.smegmatis_MC2_155 LRPVAVAALRGGVPENFRAPGVILRQVLTWVLSTGVPLLAIILALVASKF
TH_0858|M.thermoresistible__bu LRPVAVAALRGGVPENFRAPGVVLRQVLTWVLSTGVPVLAIVLALVASKV
MAB_4523|M.abscessus_ATCC_1997 LRPIAAKALSASEFHGRFAPSVFVRVRLSWWIGTLAPAIGIIALCGMALW
***:*. ** .. .. :*.* * *:* .* .* :.*:
Mb3669|M.bovis_AF2122/97 ALLHATPEALFNPILMMALAALGIG-SVSTLLVAMSIADPLRQLRWALSE
Rv3645|M.tuberculosis_H37Rv ALLHATPEALFNPILMMALAALGIG-SVSTLLVAMSIADPLRQLRWALSE
MLBr_00201|M.leprae_Br4923 ALLHAVPEQLFSPILLLALAVLVVG-LVSTWLVAMSIADPLRQLRWALSE
MMAR_5137|M.marinum_M AILHATPDKLFNPILLLALAALAIG-SASTLLVSMSIADPLRQLRWALSE
MUL_4221|M.ulcerans_Agy99 AILHATPDKLFNPILLLALAALAIG-SASTLLVSMSIADPLRQLRWALSE
MAV_0518|M.avium_104 SLLHATPEKLFTPILLLAVAALGIG-LVSTLLVAMSIADPLRQLRWALSE
Mflv_1395|M.gilvum_PYR-GCK EVLTAPADRLITTILLLAIVALIIG-LSSTVLVAMSIADPLRQLRWALGE
Mvan_5395|M.vanbaalenii_PYR-1 EVLTAPADRLITTILLLSIVALVIG-LSSTVLVAMSIADPLRQLRWALGE
MSMEG_6154|M.smegmatis_MC2_155 SILTASADRLNTPILLLAVAALVIG-MSSTVLVAMSIADPLRQLRWALGE
TH_0858|M.thermoresistible__bu GILTAPAEQLTTPLLVLAIATLVIG-LSGTVLVAMSIADPLRQLRWALGE
MAB_4523|M.abscessus_ATCC_1997 KPEDLTSTRDLALTVLLIVSTTLVGSYGLALLTAGQLADPVRQLRTAISD
. ::: : . :* : *.: .:***:**** *:.:
Mb3669|M.bovis_AF2122/97 VQRGNYNAHMQIYDASELGLLQAGFNDMVRELSERQRLRDLFGRYVGEDV
Rv3645|M.tuberculosis_H37Rv VQRGNYNAHMQIYDASELGLLQAGFNDMVRELSERQRLRDLFGRYVGEDV
MLBr_00201|M.leprae_Br4923 VQRGNYNAHMQIYDASELGLLQAGFNDMVRELSERQRLRDLFGRYVGEDV
MMAR_5137|M.marinum_M VQRGNYNAHMQIYDASELGLLQAGFNDMVRDLSERQRLRDLFGRYVGEDV
MUL_4221|M.ulcerans_Agy99 VQRGNYNAHMQIYDASELGLLQAGFNDMVRDLSERQRLRDLFGRYVGEDV
MAV_0518|M.avium_104 VQRGNYNAHMQIYDASELGLLQAGFNDMVRDLSERQRLRDLFGRYVGEDV
Mflv_1395|M.gilvum_PYR-GCK VQRGNYNAHMQIYDASELGLLQAGFNDMVRDLAERQRLRDLFGRYVGEDV
Mvan_5395|M.vanbaalenii_PYR-1 VQRGNYNAHMQIYDASELGLLQAGFNDMVRDLAERQRLRDLFGRYVGEDV
MSMEG_6154|M.smegmatis_MC2_155 VQRGNYNAHMQIYDASELGLLQAGFNDMVRDLAERQRLRDLFGRYVGEDV
TH_0858|M.thermoresistible__bu VQRGNFNAHMQIYDASELGLLQAGFNDMVRDLAERQRLRDLFGRYVGEDV
MAB_4523|M.abscessus_ATCC_1997 VAQGDFNARVEVYDGSELGVLQAGFNRMMQVGEERRQLRELFGKHVGADV
* :*::**::::**.****:****** *:: **::**:***::** **
Mb3669|M.bovis_AF2122/97 ARRALERGTELGGQERDVAVLFVDLVGSTQLAATRPPAEVVQLLNEFFRV
Rv3645|M.tuberculosis_H37Rv ARRALERGTELGGQERDVAVLFVDLVGSTQLAATRPPAEVVQLLNEFFRV
MLBr_00201|M.leprae_Br4923 ARRALEHGTELGGQERDVAVLFVDLIGSTQLAETKPPTKVVHLLNEFFRV
MMAR_5137|M.marinum_M ARRAIERGTELGGQERDVAVLFVDLVGSTQLAATRPPVEVVGLLNEFFRV
MUL_4221|M.ulcerans_Agy99 ARRAIERGTELGGQERDVAVLFVDLVGSTQLAATRPPVEVVGLLNEFFRV
MAV_0518|M.avium_104 ARRALERGTELGGQERDVAVLFVDLVGSTQLAATRPPAEVVHLLNEFFRV
Mflv_1395|M.gilvum_PYR-GCK ARRALERGTELGGQERDVAVLFVDLVGSTHLAATIPASEVVSLLNEFFRV
Mvan_5395|M.vanbaalenii_PYR-1 ARRALERGTELGGQERDVAVLFVDLVGSTHLAATRPAAEVVNLLNEFFRV
MSMEG_6154|M.smegmatis_MC2_155 ARRALERGTELGGQERDVAVLFVDLVGSTHLASTIPAADVVNLLNEFFRV
TH_0858|M.thermoresistible__bu ARRALERGTELGGQERDVAVLFVDLVGSTRLAATTPPADVVNLLNEFFRV
MAB_4523|M.abscessus_ATCC_1997 ASQALQLGTNLGGENRYVAVFFVDMIGSTSMAADRDPEEVLTILNDFFRI
* :*:: **:***::* ***:***::*** :* . .*: :**:***:
Mb3669|M.bovis_AF2122/97 VVETVARHGGFVNKFQGDAALAIFGAPIEHPDGAGAALSAARELHDELIP
Rv3645|M.tuberculosis_H37Rv VVETVARHGGFVNKFQGDAALAIFGAPIEHPDGAGAALSAARELHDELIP
MLBr_00201|M.leprae_Br4923 VVDTVGRHGGFVNKFQGDAALAIFGAPIEHPDSAGAALSAARELHDELLP
MMAR_5137|M.marinum_M VVDTVARHGGFVNKFQGDAALAIFGAPIEHPDGAGAALRAARELHDELIP
MUL_4221|M.ulcerans_Agy99 VVDTVARHGGFVNKFQGDAALAIFGAPIEHPDGAGAALRAARELHDELIP
MAV_0518|M.avium_104 VVETVGKHGGFVNKFQGDAALAIFGAPIEHPDSPGGALAAARELHDALLP
Mflv_1395|M.gilvum_PYR-GCK VVDTVNRHGGFVNKFQGDAALAIFGAPIEHPDASGAALAASRELHDELIN
Mvan_5395|M.vanbaalenii_PYR-1 VVDTVNRHGGFVNKFQGDAALAIFGAPIEHPDASGAALAASRELHDELIR
MSMEG_6154|M.smegmatis_MC2_155 IVDTVNKHGGFVNKFQGDAALAIFGAPIEHPDASGGALAASRELHDELIK
TH_0858|M.thermoresistible__bu VVDTVNRHGGFVNKFQGDAALAIFGAPIEHPDACGAALAAARELHDELIA
MAB_4523|M.abscessus_ATCC_1997 VVEVVDSRGGFVNKFVGDEALSVFGAPLHRPDATTACLAAARELRDRLNE
:*:.* :******* ** **::****:.:**. ..* *:***:* *
Mb3669|M.bovis_AF2122/97 VLG-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
Rv3645|M.tuberculosis_H37Rv VLG-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
MLBr_00201|M.leprae_Br4923 VLG-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
MMAR_5137|M.marinum_M VLG-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
MUL_4221|M.ulcerans_Agy99 VLG-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
MAV_0518|M.avium_104 VIG-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
Mflv_1395|M.gilvum_PYR-GCK VLG-ETEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
Mvan_5395|M.vanbaalenii_PYR-1 VLG-ETDFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
MSMEG_6154|M.smegmatis_MC2_155 VLGTETEFGIGVSAGRAIAGHIGAKARFEYTVIGDPVNEAARLTELAKLE
TH_0858|M.thermoresistible__bu VLG-ETEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLE
MAB_4523|M.abscessus_ATCC_1997 DFPHPFEFAIGVSAGLAVAGNVGATARYEYSVIGDPVNEASRLTELAKHR
: :*.****** *:**::** **:**:*********:******* .
Mb3669|M.bovis_AF2122/97 DGHVLASAIAVSGALDAEALCWDVGEVVELRGRAAPTQLARPMNLAAPEE
Rv3645|M.tuberculosis_H37Rv DGHVLASAIAVSGALDAEALCWDVGEVVELRGRAAPTQLARPMNLAAPEE
MLBr_00201|M.leprae_Br4923 DGHVLASAIAVSGALDAEALCWNVGEIVELRGRTAPTQLARPLNLAVPEQ
MMAR_5137|M.marinum_M EGHVLASAIAISGALDAEALCWDVAEVVELRGRTAPTQLARPLNLAVAQQ
MUL_4221|M.ulcerans_Agy99 EGHVLASAFAISGALDAEALCWDVAEVVELRGRTAPTQLARPLNLAVAQK
MAV_0518|M.avium_104 PGHVLASAIAVSGALDSEALCWDVGEVVELRGRTAPTQLARPVKLATPEE
Mflv_1395|M.gilvum_PYR-GCK PGHVLASSLAVADALDAEALCWNVGEVVELRGRTAPTQLARPVNLIRPPR
Mvan_5395|M.vanbaalenii_PYR-1 PGHVLASAIAVADALDAEALCWDVGEIVELRGRTAPTQLARPVNLVSPDD
MSMEG_6154|M.smegmatis_MC2_155 EGHVLASAIAVSGALDSEALCWDVGEIVELRGRTVPTQLARPINLARPED
TH_0858|M.thermoresistible__bu EGHVLASAMAVSGALDAEALCWDVGETVELRGRTVPTQLARPVKLAFSNG
MAB_4523|M.abscessus_ATCC_1997 PSRLLASTNALYFADPDEQKHWEHGDSVLLRGRTRATHLAWPAGT-----
.::***: *: * * *: .: * ****: .*:** *
Mb3669|M.bovis_AF2122/97 VS-----SEVRG-----------------------------
Rv3645|M.tuberculosis_H37Rv VS-----SEVRG-----------------------------
MLBr_00201|M.leprae_Br4923 IT-----SEVTG-----------------------------
MMAR_5137|M.marinum_M VS-----TKSGS-----------------------------
MUL_4221|M.ulcerans_Agy99 VS-----TKSGS-----------------------------
MAV_0518|M.avium_104 VAPQEVSSEVSG-----------------------------
Mflv_1395|M.gilvum_PYR-GCK VEE-KVADDVPSTTR--------------------------
Mvan_5395|M.vanbaalenii_PYR-1 VSQ-EVASDRR------------------------------
MSMEG_6154|M.smegmatis_MC2_155 LDG-SANGQSTDGSPAETAFQQEKGLFAVEGVEVVGASVPG
TH_0858|M.thermoresistible__bu PGP-SNNSDAPGTRTTGSVSRADAETTAPES----------
MAB_4523|M.abscessus_ATCC_1997 -----------------------------------------