For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVHPLRQRIRWLALHGVIRGLSKLAARRSGDPQARLIADPEARNDPAAFADEVRTGDPVVRCRAVLLTVD HQAANEILRSDDFRVSSLGAGLPAPLRWVNRKTHPGLLHPIEPPSLLSIEPPDHTRCRKLVSSVFTTRAV NALRDRVQLTADQLLDDLERESGVVDIVERYCSQLPVAVISDILGVPEQDRKHILHFGELGAPSLDIGLS WRQFQQVEEGLVGFNAWLGDHLEALRRNPGDDLMSELILASAAADEAGRLSERELQATAGLVLAAGFETT VNLLGNGIRMLLDTPQHLETLAARPDLWPNTVEEILRLDSPVQMSARFARRDVEVAGTRIERGELVIVHL AGANRDPNVFDDPHRFDIERDNAGRHLSFSGGRHFCLGAALARAEGEVGLRTFFQRYPDAQLAGSGSRRD TRVLRGWSTLPITLGKAPAALGS*
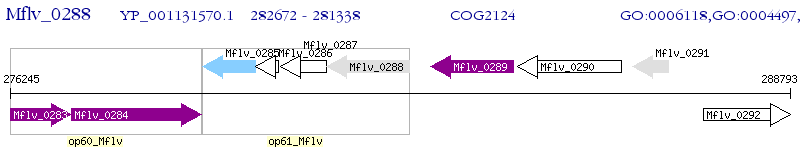
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0288 | - | - | 100% (444) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_1229 | - | 1e-60 | 38.80% (415) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_1321 | - | 1e-43 | 32.76% (409) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1912c | cyp140 | 1e-169 | 66.97% (433) | cytochrome p450 140 CYP140 |
| M. tuberculosis H37Rv | Rv1880c | cyp140 | 1e-169 | 66.97% (433) | cytochrome p450 140 CYP140 |
| M. leprae Br4923 | MLBr_02088 | - | 2e-55 | 39.21% (329) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_4456 | - | 7e-93 | 43.22% (435) | putative cytochrome P450 |
| M. marinum M | MMAR_2768 | cyp140A5 | 1e-167 | 64.69% (439) | cytochrome P450 140A5 Cyp140A5 |
| M. avium 104 | MAV_2822 | - | 1e-167 | 66.28% (433) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_0681 | - | 0.0 | 76.20% (416) | P450 heme-thiolate protein |
| M. thermoresistible (build 8) | TH_2999 | - | 0.0 | 73.73% (434) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_2979 | cyp140A5 | 1e-167 | 64.46% (439) | cytochrome P450 140A5 Cyp140A5 |
| M. vanbaalenii PYR-1 | Mvan_0600 | - | 0.0 | 87.02% (439) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1912c|M.bovis_AF2122/97 ------MKDKLHWLAMHGVIRGIAAIGIRRG-DLQARLIAD----PAVAT
Rv1880c|M.tuberculosis_H37Rv ------MKDKLHWLAMHGVIRGIAAIGIRRG-DLQARLIAD----PAVAT
MMAR_2768|M.marinum_M ------MKDKLHWFAMHGVIRGMANIGLRRG-ELEARLIAD----PAVAA
MUL_2979|M.ulcerans_Agy99 ------MKDKLHWFAMHGVIRGMANIGLRRG-ELEARLIAD----PAVAA
MAV_2822|M.avium_104 ------MKERLHWFAMHGFIRGAAALGARRG-DVHARLIAD----PAVAA
Mflv_0288|M.gilvum_PYR-GCK -MAVHPLRQRIRWLALHGVIRGLSKLAARRSGDPQARLIAD----PEARN
Mvan_0600|M.vanbaalenii_PYR-1 -MAAD-LQQRIRWLALHGVIRGLSKVAVRRGQDPQARLIAD----PAVRA
TH_2999|M.thermoresistible__bu -------------LSLHGVIRALTAYGARRG-DPQARLIAD----PAVRA
MSMEG_0681|M.smegmatis_MC2_155 ----------------------MATLGLRRG-DPQARLIAD----PVVRA
MAB_4456|M.abscessus_ATCC_1997 MRTRDQIKYWSRWATMHGISRAALLTQVRTL--PLAALFLG----PDRAE
MLBr_02088|M.leprae_Br4923 --MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRA
. * :: *
Mb1912c|M.bovis_AF2122/97 DPVPFYDEVRSHGALVRNRAN-YLTVDHRLAHDLLRSDDFRVVSFGENLP
Rv1880c|M.tuberculosis_H37Rv DPVPFYDEVRSHGALVRNRAN-YLTVDHRLAHDLLRSDDFRVVSFGENLP
MMAR_2768|M.marinum_M NPGPFHDELRRHGRMVRTRVN-YLTVDYQLAHDVLRSDDFHTISFGENLP
MUL_2979|M.ulcerans_Agy99 NPGPFHDELRRHGRMVRTRVN-YLTVDYQLAHDVLRSDDFHTISFGENLP
MAV_2822|M.avium_104 DPARFYDEARARGTLVKGRVA-YLTADHALAHELLRSEDFRVLVFGSNLP
Mflv_0288|M.gilvum_PYR-GCK DPAAFADEVRTGDPVVRCRAV-LLTVDHQAANEILRSDDFRVSSLGAGLP
Mvan_0600|M.vanbaalenii_PYR-1 DPAAFADELRGRGPVIRCRAV-LMTVDHQVVNEILRSDDFRVSSLGAGLP
TH_2999|M.thermoresistible__bu DPAAFADELRGRGPLIRGRAA-LLTFDHGVANELLRSDDFRVSELGGNLP
MSMEG_0681|M.smegmatis_MC2_155 DPTTFADQMRKQGPVIRGRVV-LLTFDHAVAFDLLRSDDFQVSQLGANLP
MAB_4456|M.abscessus_ATCC_1997 KHYRYIEEIRASGSVTPSRAGGLIFTDLALTREILRDNRFITMAP-TNIP
MLBr_02088|M.leprae_Br4923 DPFPVYRALIDYGPMQLPGMPLTVFSSFSDCDEALR----HPLSASDRLK
. . : : . : ** :
Mb1912c|M.bovis_AF2122/97 PPLR------WLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS
Rv1880c|M.tuberculosis_H37Rv PPLR------WLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS
MMAR_2768|M.marinum_M GPLR------WLERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTS
MUL_2979|M.ulcerans_Agy99 GPLR------WLERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTS
MAV_2822|M.avium_104 APLR------WLERRTRDDLLHPLRAPSLLAVEPPEHTRYRKTVSAVFTP
Mflv_0288|M.gilvum_PYR-GCK APLR------WVNRKTHPGLLHPIEPPSLLSIEPPDHTRCRKLVSSVFTT
Mvan_0600|M.vanbaalenii_PYR-1 KPLQ------WINRKTHPGLLHPIEPPSLLSIEPPDHTRCRKLVSSVFTT
TH_2999|M.thermoresistible__bu APLR------WVAAKADTGHLHPIQPPSLLSIEPPDHTRLRKLVSSVFTT
MSMEG_0681|M.smegmatis_MC2_155 KPLR------WIADKTNPGLLHPIEPPSLLSIEPPDHTRMRKLVSSVFTT
MAB_4456|M.abscessus_ATCC_1997 SPILPLAFSRWVFARTEPGLPNPVEPPAMLAVDPPEHTRFRKLVSKAFTP
MLBr_02088|M.leprae_Br4923 ATLA-------QQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAP
.: : .*. .::: ::**:*** ** ** .*:.
Mb1912c|M.bovis_AF2122/97 RAVSALRDLVEQTAINLLDRFAEQ--PGIVDVVGRYCSQLPIVVISEILG
Rv1880c|M.tuberculosis_H37Rv RAVSALRDLVEQTAINLLDRFAEQ--PGIVDVVGRYCSQLPIVVISEILG
MMAR_2768|M.marinum_M RAVAALRDRVEQTANSLLDQLAQR--PGVVDVVGQYCSQLPIMVISEILG
MUL_2979|M.ulcerans_Agy99 RAVAALRDRVEQTANSLLDQLAQR--PGVVDVVGQYCSQLPIMVISEILG
MAV_2822|M.avium_104 RAVAALRDRVERTAAELLDQLTGG--PGVVDIVGRYCSQLPVAIISEILG
Mflv_0288|M.gilvum_PYR-GCK RAVNALRDRVQLTADQLLDDLERE--SGVVDIVERYCSQLPVAVISDILG
Mvan_0600|M.vanbaalenii_PYR-1 RAVAALRERVQQTANDLLDDLERE--SGVVDIVERYCSQLPVAVISDILG
TH_2999|M.thermoresistible__bu RAVATLKERLQQTAFDLLDGLDEVGASDVVDVIDRYCSQLPIAVISDILG
MSMEG_0681|M.smegmatis_MC2_155 RAVATLRERVEQTAHELLDGLEAE--SGVVDIVERYCAQLPVAVIGDILG
MAB_4456|M.abscessus_ATCC_1997 RAVSKLEDRVREVTTELLDNLERH---ERADLLHDYASQLPVAIIAEMLG
MLBr_02088|M.leprae_Br4923 KVVQALEGDIAALVDSLLDKGAAA---GQFDVIADLAFPLAVAVICRLLG
:.* *. : . .*** *:: . *.: :* :**
Mb1912c|M.bovis_AF2122/97 VPEHDRPRVLEFGELAAPSLDIGIPWR-QYLRVQQGIRGFDCWLEGHLQQ
Rv1880c|M.tuberculosis_H37Rv VPEHDRPRVLEFGELAAPSLDIGIPWR-QYLRVQQGIRGFDCWLEGHLQQ
MMAR_2768|M.marinum_M VPEQDRGRVLEYGELAAPSLDIGVPYR-QYRRIQQGITGFNSWLTAHLEQ
MUL_2979|M.ulcerans_Agy99 VPEQDRGRVLEYGELAAPSLDIGVTYR-QYRRIQQGITGFNSWLTAHLEQ
MAV_2822|M.avium_104 VPEQDRSRVLEFGELAAPSLDIGLPWR-QYRSVQRGIAGFSSWLAGHLQQ
Mflv_0288|M.gilvum_PYR-GCK VPEQDRKHILHFGELGAPSLDIGLSWR-QFQQVEEGLVGFNAWLGDHLEA
Mvan_0600|M.vanbaalenii_PYR-1 VPERDRRHILHFGELGAPSLDIGLSWR-QYTQVHEGLVGFNEWLGGHMDE
TH_2999|M.thermoresistible__bu VPDEDRDRILHFGELGAPSLDIGLSWR-QYRQVREGIVGFNTWLTAHLQR
MSMEG_0681|M.smegmatis_MC2_155 VPEPDRPNILRYGELGAPSLDIGLSWS-QYQQVNEGIIGFETWLNRHLRT
MAB_4456|M.abscessus_ATCC_1997 VPRADAPFLLEWGNHGAALLDIGMTWS-AYRDATQALIEIDRYFDAHLVR
MLBr_02088|M.leprae_Br4923 VPYEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQ
** * . . . . :* :. . :: ::
Mb1912c|M.bovis_AF2122/97 LRHA----PGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFETT
Rv1880c|M.tuberculosis_H37Rv LRHA----PGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFETT
MMAR_2768|M.marinum_M LRRN----PGDDLMSQLIRVAESGDAGTYLDESELRAVAGLVLVAGFETT
MUL_2979|M.ulcerans_Agy99 LRRN----PGDDLMSQLIRVAESGDAGTYLDKSELRAIAGLVLVAGFETT
MAV_2822|M.avium_104 LRSN----PSDNLMSQLIQTAESGSAETYLDETELAAIAGLVLAAGFETT
Mflv_0288|M.gilvum_PYR-GCK LRRN----PGDDLMSELILASAAADEAGRLSERELQATAGLVLAAGFETT
Mvan_0600|M.vanbaalenii_PYR-1 LRRN----PGDDLMSQLIQASQAADEGSRLTDRELQATAGLVLAAGFETT
TH_2999|M.thermoresistible__bu LRRD----PGEDLLSQLIQAADAET-----TERELEALAGLVLAAGFETT
MSMEG_0681|M.smegmatis_MC2_155 LRRD----PGDDLLSQIITASESGATGEPLTDRELKALAGLVLAAGFETT
MAB_4456|M.abscessus_ATCC_1997 LRGE----LAEDPTVDGILASIVRDGD--LNDRELKATMALLLGAGFETT
MLBr_02088|M.leprae_Br4923 LVKCRRGTPGEDLISRLIELDESGDQ---LTEEEIIATCGLLLVAGHETT
* .:: * . *: * .*:* **.***
Mb1912c|M.bovis_AF2122/97 VNLLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVAC
Rv1880c|M.tuberculosis_H37Rv VNLLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVAC
MMAR_2768|M.marinum_M VNLLGNGIRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQLTARVAR
MUL_2979|M.ulcerans_Agy99 VNLLGNGIRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQLTARVAR
MAV_2822|M.avium_104 VNLLGNGIRMLLDAPEHLDTLRRRPELWPNAVEEILRLESPVQLTARMAL
Mflv_0288|M.gilvum_PYR-GCK VNLLGNGIRMLLDTPQHLETLAARPDLWPNTVEEILRLDSPVQMSARFAR
Mvan_0600|M.vanbaalenii_PYR-1 VNLLGNGIRMLLETPGHLETLAARPELWPNAVEEILRLDSPVQMSARFAR
TH_2999|M.thermoresistible__bu VNLLGNGIRMLLDTPGHLDTLAARPELWPNAVEEILRLDPPVQMTARVAR
MSMEG_0681|M.smegmatis_MC2_155 VNLLGNGIRMLLDHREHLETLAARPDVWPNAVEEILRLDSPVQMTARAAV
MAB_4456|M.abscessus_ATCC_1997 VNLIGNGIAALLRHPQQLAHLRENPDGWPNAVEEILRYDSPVQITGRVAT
MLBr_02088|M.leprae_Br4923 VNLIANAVLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAA
***:.*.: :* : * .*: * .*** ** :..::: .* *
Mb1912c|M.bovis_AF2122/97 RDVEVAGVRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERPNAGRHLAFS
Rv1880c|M.tuberculosis_H37Rv RDVEVAGVRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERPNAGRHLAFS
MMAR_2768|M.marinum_M TDVEIASWPINRGEMVVAYLAGANRDPAVFPNPHRFDVERANAGRHLAFS
MUL_2979|M.ulcerans_Agy99 TDVEIASWPINRGEMVVAYLAGANRDPAVFPNPHRFDVERANAGRHLAFS
MAV_2822|M.avium_104 NDVEVAGRQLHRGDLVLVYLAAANRDPAVFGDPHRFDIERPNAGRHLAFS
Mflv_0288|M.gilvum_PYR-GCK RDVEVAGTRIERGELVIVHLAGANRDPNVFDDPHRFDIERDNAGRHLSFS
Mvan_0600|M.vanbaalenii_PYR-1 RDVTVGGTEIKRGELVILHLAGANRDPEVFTDPHRFDIERDNAGRHLSFS
TH_2999|M.thermoresistible__bu VDTDIAGTAVRRGEMVIIHLAGANRDPKVFADPHRFDIERANAHRHLSFS
MSMEG_0681|M.smegmatis_MC2_155 RDVDVAGTAVGRGELVVLHIAGANRDPKVFDDPHRFDIERDNAGRHLSFS
MAB_4456|M.abscessus_ATCC_1997 QACEFEGHTLAAGSMAILLLGGANRDPAVFDQPDVFDVLRANAREHVAFG
MLBr_02088|M.leprae_Br4923 KDMTIGQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPSS-RHLAFA
. : *. : :..***** *: *. ** * .: .*::*.
Mb1912c|M.bovis_AF2122/97 TGRHFCLGAALARAEGEVGLRTFFDRFPDVRAAGAGSRRDTRVLRGWSTL
Rv1880c|M.tuberculosis_H37Rv TGRHFCLGAALARAEGEVGLRTFFDRFPDVRAAGAGSRRDTRVLRGWSTL
MMAR_2768|M.marinum_M TGRHFCLGAALARAEGEVGLRTFFERFPDVRAAGAGSRRDTRVLRGWSTL
MUL_2979|M.ulcerans_Agy99 TGRHFCLGAALARAEGEVGLRTFFERFPDVRAAGAGSRRDTRVLRGWSTL
MAV_2822|M.avium_104 GGRHFCLGAALARAEGEVGLRTFFERFPEARAAGAGSRRETRVLRGWSSL
Mflv_0288|M.gilvum_PYR-GCK GGRHFCLGAALARAEGEVGLRTFFQRYPDAQLAGSGSRRDTRVLRGWSTL
Mvan_0600|M.vanbaalenii_PYR-1 GGRHFCLGAALARAEGEVGLRTFFERYPDARLAGQGSRRDTRVLRGWSTL
TH_2999|M.thermoresistible__bu GGRHFCLGAALARAEGEVGLRTFFQRYPKARSAGGGDRRDTRVLRGWASL
MSMEG_0681|M.smegmatis_MC2_155 GGRHFCLGAALARAEGEVGLRAFFDRFPDARLAGDPVRRDTRVLRGWSTL
MAB_4456|M.abscessus_ATCC_1997 SGIHVCLGASLARMEGVVALQSLFERFPELALAADPTPGKHVNLHGFGSL
MLBr_02088|M.leprae_Br4923 VGSHFCLGAALARLEATVTLSAISARFPQVQLAGELVYKPNVAMRGMSAL
* *.****:*** *. * * :: *:*. *. ::* .:*
Mb1912c|M.bovis_AF2122/97 PVTLGPARSMVSP------------
Rv1880c|M.tuberculosis_H37Rv PVTLGPARSMVSP------------
MMAR_2768|M.marinum_M PVTLGPARALAAS------------
MUL_2979|M.ulcerans_Agy99 PVTLGPARALAAS------------
MAV_2822|M.avium_104 PVRLGPARSLAAAEAGGRPDEPTAG
Mflv_0288|M.gilvum_PYR-GCK PITLGKAPAALGS------------
Mvan_0600|M.vanbaalenii_PYR-1 PITLGTARAALGS------------
TH_2999|M.thermoresistible__bu PITLGPARTTVGS------------
MSMEG_0681|M.smegmatis_MC2_155 PIALGKARAAVGS------------
MAB_4456|M.abscessus_ATCC_1997 PVNLGRARVPASS------------
MLBr_02088|M.leprae_Br4923 PVQV---------------------
*: :