For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSKMSDIQEKDPVRTGIFGIAVVSMLVLVAFGYTGLPFWPQGKQYTGYFADSGGLESGGNVFVSGIKVGQ VKDVSLAGNKVRVDFTVDRSIRLGDQSLAAIRTTTVLGEKSLSVTPAGAGRITEIPLGRTTTPYTLNSAL SDLGASAGGIDKAQLDQSLQVITEAMRDATPQLRGALDGVASLSRTLNARDAALEQLLTHAKSVSGILSE RAGQVNKLVTDGNQLFAALDERRLAISELINRIQGLSQQISGFVADNRQEFRPTMEKLSVVLDDLLDRKA HISEALKRLPGYGTALGEVVSSGPGFHVNVYGFPPATLTAVAFDTYFQPGKLPASLSDYLNGLIGERQIM RPKSP*
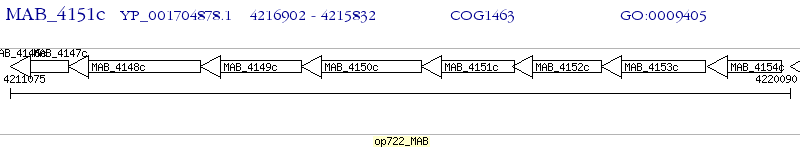
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4151c | - | - | 100% (356) | MCE-family protein |
| M. abscessus ATCC 19977 | MAB_4597c | - | 6e-30 | 29.43% (282) | putative Mce family protein |
| M. abscessus ATCC 19977 | MAB_1697 | - | 1e-24 | 26.00% (300) | putative Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3527c | mce4C | 1e-127 | 63.46% (353) | MCE-family protein MCE4C |
| M. gilvum PYR-GCK | Mflv_1558 | - | 1e-129 | 63.92% (352) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv3497c | mce4C | 1e-127 | 63.46% (353) | MCE-family protein MCE4C |
| M. leprae Br4923 | MLBr_02591 | mce1C | 6e-44 | 30.79% (328) | putative secreted protein |
| M. marinum M | MMAR_4985 | mce4C | 1e-128 | 63.82% (351) | MCE-family protein Mce4C |
| M. avium 104 | MAV_0659 | - | 1e-130 | 64.66% (348) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_5898 | - | 1e-132 | 64.77% (352) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_1935 | - | 1e-124 | 61.36% (352) | MCE-FAMILY PROTEIN MCE4C |
| M. ulcerans Agy99 | MUL_4060 | mce4C | 1e-126 | 62.86% (350) | MCE-family protein Mce4C |
| M. vanbaalenii PYR-1 | Mvan_5200 | - | 1e-133 | 64.59% (353) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3527c|M.bovis_AF2122/97 MLNRKPSSKHERDPLRTGIFGLVLVICVVLIAFGYSGLPFWPQGKTYDAY
Rv3497c|M.tuberculosis_H37Rv MLNRKPSSKHERDPLRTGIFGLVLVICVVLIAFGYSGLPFWPQGKTYDAY
MMAR_4985|M.marinum_M ----MPSSKHDRDPLRTGIFGVVLVICVVLIAFGYSGLPFLPQGKTYNAY
MUL_4060|M.ulcerans_Agy99 ----MPSSKHDRDSLRTGIFGVVLVICVVLIAFGYSGLPFLPQGKTYNAY
MAV_0659|M.avium_104 ---MSSNAPRERDPLRTGIFGVVLVVCVVLIAFGYAGLPFWPQGKIYDAY
Mflv_1558|M.gilvum_PYR-GCK -----MARPDDSNPLRTGIFGIALVACLVLVSFGYSSLPFFPQGKAYEAY
Mvan_5200|M.vanbaalenii_PYR-1 --MPKLSSSEDSNPLRTGIFGIALVTCLVLVSFGYTSLPFWPQGKSYEAY
MSMEG_5898|M.smegmatis_MC2_155 -----MASLRDDKTLRTGIFGIVLVTAVVLVSFGYTGLPFFPQGKPYEAY
TH_1935|M.thermoresistible__bu -----MADTGERNPLRTGIFGIAIVMCVVLVAFGFQTLPFLKQGRAYAAY
MAB_4151c|M.abscessus_ATCC_199 -MPSKMSDIQEKDPVRTGIFGIAVVSMLVLVAFGYTGLPFWPQGKQYTGY
MLBr_02591|M.leprae_Br4923 -----MRTLELPNRLRSGLIGVLVVLLIIGVGQSFTSVPILFARPSYYGQ
. :*:*::*: :* :: :. .: :*: * .
Mb3527c|M.bovis_AF2122/97 FTDAGGITPGNSVYVSGLKVGAVSAVSLAGNSAKVTFSVDRSIVVGDQSL
Rv3497c|M.tuberculosis_H37Rv FTDAGGITPGNSVYVSGLKVGAVSAVSLAGNSAKVTFSVDRSIVVGDQSL
MMAR_4985|M.marinum_M FSDAGGITPGNNVYVSGFKVGTVSDVSLAGDAAKITFSVDRHVVVGDQSL
MUL_4060|M.ulcerans_Agy99 FSDAGGITPGNNVYVSGFKVGTVSDVSLAGDAAKITFSVDRHVVVGDQSL
MAV_0659|M.avium_104 FTDAGGINPGNAVYVSGLKVGKVTDVGLAGDSAKISFSVDRHVAVGDQSL
Mflv_1558|M.gilvum_PYR-GCK FADAGGITPGNDVNVSGINVGKVNSVELAGDAAKITFTVNREVKVGDQSL
Mvan_5200|M.vanbaalenii_PYR-1 FADAGGITPGNSVNVSGISVGKVSAVELAGDAAKITFTVDREVKVGDQSL
MSMEG_5898|M.smegmatis_MC2_155 FADAGGIEVGNDVNVSGITVGKVSDVELAGDAAKVTFTVDRKIKVGDQSL
TH_1935|M.thermoresistible__bu FADAGGITPGNAVNISGIKVGDVTGVHLAGDKAKVTFTVDRTVRVGDQSL
MAB_4151c|M.abscessus_ATCC_199 FADSGGLESGGNVFVSGIKVGQVKDVSLAGNKVRVDFTVDRSIRLGDQSL
MLBr_02591|M.leprae_Br4923 FTDTGGLNKGDKVRIAGMDVGKVEALKIDGDHVVIKFSIG-TNRIGTASR
*:*:**: *. * ::*: ** * : : *: . : *::. :* *
Mb3527c|M.bovis_AF2122/97 AAIRTDTILGERSIAVSPAGSGKS---TTIPLSRTTTPYTLNGVLQDLGR
Rv3497c|M.tuberculosis_H37Rv AAIRTDTILGERSIAVSPAGSGKS---TTIPLSRTTTPYTLNGVLQDLGR
MMAR_4985|M.marinum_M AAIRTDTILGERSVAVTPAGSGNA---TTIPLSRTTTPYTLNGALQDLGG
MUL_4060|M.ulcerans_Agy99 AAIRTDTILGERSVAVTPAGSGNA---TTIPLSRTTTPYTLNGALKDLGG
MAV_0659|M.avium_104 AAIRTDTILGERSIAVTPAGGGKA---TTIPLSRTTTPYTLAGALEDLGS
Mflv_1558|M.gilvum_PYR-GCK VAIKTDTVLGEKSLSVTPKGNGES---TVIPLGRTTTPYTLNTALQDLGQ
Mvan_5200|M.vanbaalenii_PYR-1 VAIKTDTVLGEKSLAITPKGSGES---TVIPLGRTTTPYTLNTALQDLGQ
MSMEG_5898|M.smegmatis_MC2_155 VAIKTDTVLGQKSLSVTPKGAGSP---SVIPLGRTTTPYTLNTALQDLGQ
TH_1935|M.thermoresistible__bu ASIKTDTVLGEKSLALKPAGSGQV---TVIPAERTTSPYTLNNALQDLTT
MAB_4151c|M.abscessus_ATCC_199 AAIRTTTVLGEKSLSVTPAGAGRI---TEIPLGRTTTPYTLNSALSDLGA
MLBr_02591|M.leprae_Br4923 LGIRTDTVLGKKVLEVETRGTQLLRPGDSLPLGQSTTPYQSYDAFFDATK
.*:* *:**:: : : . * :* ::*:** .: *
Mb3527c|M.bovis_AF2122/97 NANDLNRPQFEQALNVFTQALHDATPQVRGAVDGLTSLSRALNRRDEALQ
Rv3497c|M.tuberculosis_H37Rv NANDLNRPQFEQALNVFTQALHDATPQVRGAVDGLTSLSRALNRRDEALQ
MMAR_4985|M.marinum_M SASNLDKPQFEHALQVLTETLHDATPQLRGALDGVTALSRTLNTRDEALQ
MUL_4060|M.ulcerans_Agy99 SASNLDKPQFEHALQVLTETLHDATPQLRGALDGVTALSRTLNTRDEALQ
MAV_0659|M.avium_104 NASNLNKPQFEQALHVLTDTLHDATPELRGALDGVTSLSRALDRRDEALQ
Mflv_1558|M.gilvum_PYR-GCK NVGELDKPQFEMALQTLTDSLKDATPHLRSALDGVTNLSRSLNKRDEALD
Mvan_5200|M.vanbaalenii_PYR-1 NVSALDKPRFEMALQTLTDSLKDATPQLRNALDGVANLSRSLNARDEALD
MSMEG_5898|M.smegmatis_MC2_155 NVGELDKPRFEQALQTLTDTLRDATPQLRSALDGVANLSRSINRRDEALG
TH_1935|M.thermoresistible__bu NVGDLDQERFTEALGLLTEALQDATPHLRSTLDGVTALSASINANDAAIA
MAB_4151c|M.abscessus_ATCC_199 SAGGIDKAQLDQSLQVITEAMRDATPQLRGALDGVASLSRTLNARDAALE
MLBr_02591|M.leprae_Br4923 VASGWNIDTIKQSLKVVSETIDQTYPHLSAALDGVAKFSDTIGARDKEIK
.. : : :* .:::: :: *.: ::**:: :* ::. .* :
Mb3527c|M.bovis_AF2122/97 GLLAHAKSVTSVLSERAEQVNKLVEDGNQLFAALDARRAALSALISGIDD
Rv3497c|M.tuberculosis_H37Rv GLLAHAKSVTSVLSERAEQVNKLVEDGNQLFAALDARRAALSALISGIDD
MMAR_4985|M.marinum_M GLLAHAKTVTGVLSERAAQVNKLIQDGNQLFAALDERRSALGALISGIDD
MUL_4060|M.ulcerans_Agy99 GLLAHAKTVTGVLSERAAQVDKLIQDGNQLFAALDERRSALGALISGIDD
MAV_0659|M.avium_104 SLLVHAKSVTGVLSQRAEQVNKLVDDGNELFAALDERRAALGRLISGIQG
Mflv_1558|M.gilvum_PYR-GCK QLLARAKNVSDTLAQRSGQVNQLITDGNLLFAALDERRQALSNLIAGIDD
Mvan_5200|M.vanbaalenii_PYR-1 QLLARAKNVSDTLAQRSGQVNQLITDGNLLFAALDERRQALSNLIAGIDD
MSMEG_5898|M.smegmatis_MC2_155 QLLEHAKRVSDLLAQRAGQVNQLITDGNQLFAALDARRQALSTLIDGIDD
TH_1935|M.thermoresistible__bu QLLARARSVSQVLAERAGQVNQLVLDGNRLLAALDERRAALSALIAGIDD
MAB_4151c|M.abscessus_ATCC_199 QLLTHAKSVSGILSERAGQVNKLVTDGNQLFAALDERRLAISELINRIQG
MLBr_02591|M.leprae_Br4923 HLIAQANQVASVLGDRGAQVDRLLVNTKTLIAAFNERGRAVDALLGNVAA
*: :*. *: *.:*. **::*: : : *:**:: * *:. *: :
Mb3527c|M.bovis_AF2122/97 VAAQISGFVADNRKEFGPALSKLNLVLANLNERRDYITEALKRLPTYATT
Rv3497c|M.tuberculosis_H37Rv VAAQISGFVADNRKEFGPALSKLNLVLANLNERRDYITEALKRLPTYATT
MMAR_4985|M.marinum_M VSVQLSGFVNDNRKEFGPALSKLNLVVSNLNERRDYITEALKRLPAYATT
MUL_4060|M.ulcerans_Agy99 VSVQLSGFVNDNRKEFGRALSKLNLVVSNLNERRDYITEALKRLPAYATT
MAV_0659|M.avium_104 LSAQISGFVADNRKEFGPALNKLNDVLANLNERRDYITEALKRLPTYATE
Mflv_1558|M.gilvum_PYR-GCK VATQISGFVADNRREFKPALEKLNLVLDNLLERREHIGEALKRLPAYATA
Mvan_5200|M.vanbaalenii_PYR-1 VATQISGFVADNKREFKPALEKLNLVLDNLLERREHISEALKRLPAYATA
MSMEG_5898|M.smegmatis_MC2_155 VSRQLSGFVADNRREFKPALDKLNLVMDNLLERREHIGEALRRLPPYATA
TH_1935|M.thermoresistible__bu VSEQLSGFVADNRREFGPALAKLNLVLDNLLERREHISAALKRGPAYATA
MAB_4151c|M.abscessus_ATCC_199 LSQQISGFVADNRQEFRPTMEKLSVVLDDLLDRKAHISEALKRLPGYGTA
MLBr_02591|M.leprae_Br4923 FAAQVQRLINDN-PNLNHVLEQLHQLSGILVQHKDDLANTLIQVAAFLPS
.: *:. :: ** :: .: :* : * ::: : :* : . : .
Mb3527c|M.bovis_AF2122/97 LGEVVGSGPGFNVNVYSVLPGPLVATVFDLVFQ-----------------
Rv3497c|M.tuberculosis_H37Rv LGEVVGSGPGFNVNVYSVLPGPLVATVFDLVFQ-----------------
MMAR_4985|M.marinum_M LGEVVGSGPGFNVNVNSVLPAPMVAMVFDTVFQ-----------------
MUL_4060|M.ulcerans_Agy99 LGEVVGSGPGFNVNVYSVLPAPMVAMVFDTVFQ-----------------
MAV_0659|M.avium_104 LGEVVGSGPGFNVNVYGVIPAPLLATMFDFFYQ-----------------
Mflv_1558|M.gilvum_PYR-GCK LGEVVGSGPGFQINLYGLPPAPIAEVLLDTYFQ-----------------
Mvan_5200|M.vanbaalenii_PYR-1 LGEVVGSGPGFQINLYGLPPAPIAEVLLDTYFQ-----------------
MSMEG_5898|M.smegmatis_MC2_155 LGEVVGSGPGFQINLYGLPPAAISEVLLDTYFQ-----------------
TH_1935|M.thermoresistible__bu LGEVVGSGPGFHINLYGLPPGPMAEVLLDVYFQ-----------------
MAB_4151c|M.abscessus_ATCC_199 LGEVVSSGPGFHVNVYGFPPATLTAVAFDTYFQ-----------------
MLBr_02591|M.leprae_Br4923 LNEAIGSGPFFKVVLHNLAPYQILQPWVDAAFKKRGIDPENFWRSAGLPE
*.*.:.*** *:: : .. * : .* ::
Mb3527c|M.bovis_AF2122/97 -----------PGKLPDSLADYLRGFIQ----------------------
Rv3497c|M.tuberculosis_H37Rv -----------PGKLPDSLADYLRGFIQ----------------------
MMAR_4985|M.marinum_M -----------PGKLPDSFADYLRGLIQ----------------------
MUL_4060|M.ulcerans_Agy99 -----------PGKLPDSFADYLRGLIQ----------------------
MAV_0659|M.avium_104 -----------PGKMPASLADYLRGLIQ----------------------
Mflv_1558|M.gilvum_PYR-GCK -----------PGKLPDSLSDMLRGYIS----------------------
Mvan_5200|M.vanbaalenii_PYR-1 -----------PGKLPDSLSDMLRGYIS----------------------
MSMEG_5898|M.smegmatis_MC2_155 -----------PGKLPDSLADMLRGYIA----------------------
TH_1935|M.thermoresistible__bu -----------PGKLPDSLADYLRGFIS----------------------
MAB_4151c|M.abscessus_ATCC_199 -----------PGKLPASLSDYLNGLIG----------------------
MLBr_02591|M.leprae_Br4923 FRFPDPNGTRFPNGAPPPAPPVLEGTPENPGPAVPPGSPCSYTPAHPGGN
*. * . . *.*
Mb3527c|M.bovis_AF2122/97 -----ERWIIRPKSP-----------------------------------
Rv3497c|M.tuberculosis_H37Rv -----ERWIIRPKSP-----------------------------------
MMAR_4985|M.marinum_M -----ERWIIRPKSP-----------------------------------
MUL_4060|M.ulcerans_Agy99 -----ERWTIRPKSS-----------------------------------
MAV_0659|M.avium_104 -----ERWIIRPKSP-----------------------------------
Mflv_1558|M.gilvum_PYR-GCK -----ERMILRPKSP-----------------------------------
Mvan_5200|M.vanbaalenii_PYR-1 -----ERMIIRPKSP-----------------------------------
MSMEG_5898|M.smegmatis_MC2_155 -----ERTIVRPKSP-----------------------------------
TH_1935|M.thermoresistible__bu -----ERLILRPKSP-----------------------------------
MAB_4151c|M.abscessus_ATCC_199 -----ERQIMRPKSP-----------------------------------
MLBr_02591|M.leprae_Br4923 NTIGDEMGLPRPWNPLPCAGATAGPYGGPAFPAPIDVQTSPPNPDGLPLL
* ** ..
Mb3527c|M.bovis_AF2122/97 --------------------------------------------------
Rv3497c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4985|M.marinum_M --------------------------------------------------
MUL_4060|M.ulcerans_Agy99 --------------------------------------------------
MAV_0659|M.avium_104 --------------------------------------------------
Mflv_1558|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5200|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5898|M.smegmatis_MC2_155 --------------------------------------------------
TH_1935|M.thermoresistible__bu --------------------------------------------------
MAB_4151c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02591|M.leprae_Br4923 PGIAIAGRPGDPAPNVPGTPVPLPPNAPPGARTEQLAPAAPAQDSSAPHE
Mb3527c|M.bovis_AF2122/97 --------------------------
Rv3497c|M.tuberculosis_H37Rv --------------------------
MMAR_4985|M.marinum_M --------------------------
MUL_4060|M.ulcerans_Agy99 --------------------------
MAV_0659|M.avium_104 --------------------------
Mflv_1558|M.gilvum_PYR-GCK --------------------------
Mvan_5200|M.vanbaalenii_PYR-1 --------------------------
MSMEG_5898|M.smegmatis_MC2_155 --------------------------
TH_1935|M.thermoresistible__bu --------------------------
MAB_4151c|M.abscessus_ATCC_199 --------------------------
MLBr_02591|M.leprae_Br4923 PREQLSAPLINPGGTGGSGGAGGSRN