For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTLELPNRLRSGLIGVLVVLLIIGVGQSFTSVPILFARPSYYGQFTDTGGLNKGDKVRIAGMDVGKVEA LKIDGDHVVIKFSIGTNRIGTASRLGIRTDTVLGKKVLEVETRGTQLLRPGDSLPLGQSTTPYQSYDAFF DATKVASGWNIDTIKQSLKVVSETIDQTYPHLSAALDGVAKFSDTIGARDKEIKHLIAQANQVASVLGDR GAQVDRLLVNTKTLIAAFNERGRAVDALLGNVAAFAAQVQRLINDNPNLNHVLEQLHQLSGILVQHKDDL ANTLIQVAAFLPSLNEAIGSGPFFKVVLHNLAPYQILQPWVDAAFKKRGIDPENFWRSAGLPEFRFPDPN GTRFPNGAPPPAPPVLEGTPENPGPAVPPGSPCSYTPAHPGGNNTIGDEMGLPRPWNPLPCAGATAGPYG GPAFPAPIDVQTSPPNPDGLPLLPGIAIAGRPGDPAPNVPGTPVPLPPNAPPGARTEQLAPAAPAQDSSA PHEPREQLSAPLINPGGTGGSGGAGGSRN
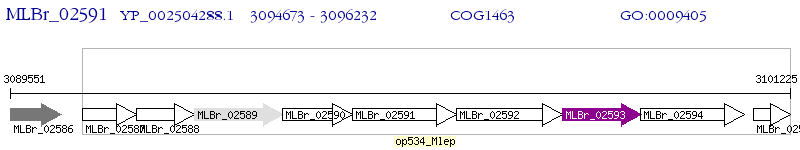
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02591 | mce1C | - | 100% (519) | putative secreted protein |
| M. leprae Br4923 | MLBr_02590 | mce1B | 8e-14 | 23.99% (296) | putative secreted protein |
| M. leprae Br4923 | MLBr_02593 | lprK | 3e-09 | 20.93% (258) | putative lipoprotein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0177 | mce1C | 0.0 | 72.38% (525) | MCE-family protein MCE1C |
| M. gilvum PYR-GCK | Mflv_0709 | - | 1e-168 | 58.99% (517) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0171 | mce1C | 0.0 | 72.38% (525) | MCE-family protein MCE1C |
| M. abscessus ATCC 19977 | MAB_4151c | - | 3e-43 | 30.79% (328) | MCE-family protein |
| M. marinum M | MMAR_0414 | mce1C | 0.0 | 72.32% (531) | MCE-family protein Mce1C |
| M. avium 104 | MAV_5013 | - | 0.0 | 75.90% (527) | mce-family protein mce1c |
| M. smegmatis MC2 155 | MSMEG_0136 | - | 1e-166 | 58.24% (510) | virulence factor Mce family protein, putative |
| M. thermoresistible (build 8) | TH_3066 | - | 1e-159 | 57.93% (492) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_1064 | mce1C | 0.0 | 71.75% (531) | MCE-family protein Mce1C |
| M. vanbaalenii PYR-1 | Mvan_0136 | - | 1e-170 | 58.49% (518) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0709|M.gilvum_PYR-GCK ----MRVLEGSNRVRGGLMGIILLVVVIGVGQSFASVPMLFATPTYYAEF
Mvan_0136|M.vanbaalenii_PYR-1 ----MRVLEGSNRIRGGMMGIIILVLVIGVGQSFASVPMLFATPTYYAQF
MSMEG_0136|M.smegmatis_MC2_155 ----MRTLQGSDRFRKGLMGVIVVALIIGVGSTLTSVPMLFAVPTYYGQF
TH_3066|M.thermoresistible__bu -------LEESDRLRNGLKGIIILLLVIGVGQSFASVPMLFATPTYYAQF
MMAR_0414|M.marinum_M ----MRTLEPPNRLRIGLMGIVVMLVVVGVGQSFTSVPMIFARPSYYGQF
MUL_1064|M.ulcerans_Agy99 ----MRTLEPPNRLRIGLMGIVVMLVVVGVGQSFTSVPMIFARPSYYGQF
MAV_5013|M.avium_104 ----MRTLEPPNRVRIGLMGIVVTVLVIGVGQSFTSVPMLFAKPSYYGQF
Mb0177|M.bovis_AF2122/97 ----MRTLEPPNRMRIGLMGIVVALLVVAVGQSFTSVPMLFAKPSYYGQF
Rv0171|M.tuberculosis_H37Rv ----MRTLEPPNRMRIGLMGIVVALLVVAVGQSFTSVPMLFAKPSYYGQF
MLBr_02591|M.leprae_Br4923 ----MRTLELPNRLRSGLIGVLVVLLIIGVGQSFTSVPILFARPSYYGQF
MAB_4151c|M.abscessus_ATCC_199 MPSKMSDIQEKDPVRTGIFGIAVVSMLVLVAFGYTGLPFWPQGKQYTGYF
:: : .* *: *: : ::: *. :.:*: * . *
Mflv_0709|M.gilvum_PYR-GCK SDTGGLNSGDNVRIAGVDVGTVRSLEIDGDRVRIGYSLG-GTEIGTDSLA
Mvan_0136|M.vanbaalenii_PYR-1 SDTGGINSGDKVRIAGVDVGTVRSMEIDGDRVLIGYGLG-GTEIGTESRV
MSMEG_0136|M.smegmatis_MC2_155 ADTGGLNIGDKVRIAGMDVGNVKSMEIDGDKVVIGYTLG-GRTIGTESRA
TH_3066|M.thermoresistible__bu YDTGGIKAGDKVRIAGVDVGLVRSATINGDKVRIGFTLG-GVQIGTDSRA
MMAR_0414|M.marinum_M TDSGGINNGDKVRIAGLDVGKVEGLRIDGDHIVMKFSIG-TNTIGTDSRL
MUL_1064|M.ulcerans_Agy99 TDSGGINNGDKVRIAGLDVGKVEGLRIDGDHIVMKFSIG-TNTIGTDSRL
MAV_5013|M.avium_104 TDSGGINTGDKVRIAGMDVGKVEGLKIDGDHIVVKFSIG-TNTIGTESRL
Mb0177|M.bovis_AF2122/97 TDSGGLHKGDRVRIAGLGVGTVEGLKIDGDHIVVKFSIG-TNTIGTESRL
Rv0171|M.tuberculosis_H37Rv TDSGGLHKGDRVRIAGLGVGTVEGLKIDGDHIVVKFSIG-TNTIGTESRL
MLBr_02591|M.leprae_Br4923 TDTGGLNKGDKVRIAGMDVGKVEALKIDGDHVVIKFSIG-TNRIGTASRL
MAB_4151c|M.abscessus_ATCC_199 ADSGGLESGGNVFVSGIKVGQVKDVSLAGNKVRVDFTVDRSIRLGDQSLA
*:**:. *..* ::*: ** *. : *::: : : :. :* *
Mflv_0709|M.gilvum_PYR-GCK AIRTDTILGRRNMEIEPRGSDPLQANGVLPLGQTTTPYQIYDAFFDVSKN
Mvan_0136|M.vanbaalenii_PYR-1 AIRTDTILGRRNLEIDPRGSEPLRANGVLPLGQTTTPYQIYDAFFDVSNN
MSMEG_0136|M.smegmatis_MC2_155 AIRTDTILGRKNIEIEPRGSETLKPRGVLPVGQTSAPYQIYDAFLDVTRN
TH_3066|M.thermoresistible__bu SIRTDTILGRKNMEIEPRGTEPLRTNGVLPIGQTTTPYQIYDAFFDVTKA
MMAR_0414|M.marinum_M SIRTDTILGKKVVEIEARGNQPLRPGGTLPLGQSTTPYQIYDAFFDVTRA
MUL_1064|M.ulcerans_Agy99 SIRTDTILGKKVVEIEARGNQPLRPGGTLPLGQSTTPYQIYDAFFDVTRA
MAV_5013|M.avium_104 AIKTDTILGKKILDVEARGSQQLRPGSTLPLGQSTTPYQIYDAFFDVTKA
Mb0177|M.bovis_AF2122/97 AIRTDTILGRKVLEIEPRGAQALPPGGVLPVGQSTTPYQIYDAFFDVTKA
Rv0171|M.tuberculosis_H37Rv AIRTDTILGRKVLEIEPRGAQALPPGGVLPVGQSTTPYQIYDAFFDVTKA
MLBr_02591|M.leprae_Br4923 GIRTDTVLGKKVLEVETRGTQLLRPGDSLPLGQSTTPYQSYDAFFDATKV
MAB_4151c|M.abscessus_ATCC_199 AIRTTTVLGEKSLSVTPAGAGRIT---EIPLGRTTTPYTLNSALSDLGAS
.*:* *:**.: :.: . * : :*:*::::** .*: *
Mflv_0709|M.gilvum_PYR-GCK TAGWDTQTVKESLNVLSETIDQTYPHLSAALDGVARFSDTIGKRDDQIKQ
Mvan_0136|M.vanbaalenii_PYR-1 TAGWDTKTVRDSLNVLSETIDQTSPHLSAALDGVARFSDTIGKRDEQIKQ
MSMEG_0136|M.smegmatis_MC2_155 AAGWDTQAVRQSLNVLSETVDQTSPHLSAALDGVARFSETIGKRDEDVKK
TH_3066|M.thermoresistible__bu ATEWDTRTVRESLNVLSETIDQTYPHLSAALDGVARFSDTIGKRDEQITQ
MMAR_0414|M.marinum_M ATGWDIDTVKKSLHVLSETIDQTYPHLSSALDGVAKFSDTLGKRDDELKH
MUL_1064|M.ulcerans_Agy99 ATGWDIDTVKKSLHVLSETIDQTYPHLSSALDGVAKFSDTLGKRDDELKH
MAV_5013|M.avium_104 AQGWDIETVKQSLHVLSQTIDQTYPHLSPALDGVAKFSDTIGKRDEQVKH
Mb0177|M.bovis_AF2122/97 ASGWDIETVKRSLNVLSETVDQTYPHLSAALDGVAKFSDTIGKRDEQITH
Rv0171|M.tuberculosis_H37Rv ASGWDIETVKRSLNVLSETVDQTYPHLSAALDGVAKFSDTIGKRDEQITH
MLBr_02591|M.leprae_Br4923 ASGWNIDTIKQSLKVVSETIDQTYPHLSAALDGVAKFSDTIGARDKEIKH
MAB_4151c|M.abscessus_ATCC_199 AGGIDKAQLDQSLQVITEAMRDATPQLRGALDGVASLSRTLNARDAALEQ
: : : **:*::::: :: *:* ****** :* *:. ** : :
Mflv_0709|M.gilvum_PYR-GCK LLANANKVAGVLGNRSEQINQLFVNAQTLLAAVNERNYAVSQLLERVDTF
Mvan_0136|M.vanbaalenii_PYR-1 LLANANKVAGVLGNRSEQINQLFVNAQSLLAAINERNYAVSQLLERVDTF
MSMEG_0136|M.smegmatis_MC2_155 LLASANKVATVLGDRSTQVNQLLVNAQTLLAAVNERGRSVSLLLERVSSV
TH_3066|M.thermoresistible__bu LLANANKIAEVLGDRGGQINALLVNAQTLLGAINERTQAVDLLLQRISQV
MMAR_0414|M.marinum_M LLAQANQVASVLGDRSAQVDRLLVNAKTLLVAFNERGRAIDALLGNIASF
MUL_1064|M.ulcerans_Agy99 LLAQANQVASALGDRSAQVDRLLVNAKTLLVAFNERGRAIDALLGNIASF
MAV_5013|M.avium_104 LLAQANQVASVLGDRSDQIDRLLVNTKTLLAAFNERGQAINALLGNIAAF
Mb0177|M.bovis_AF2122/97 LLAQANQVASILGDRSDQVDRLLVNAKTLIAAFNERGRAVDALLGNISAF
Rv0171|M.tuberculosis_H37Rv LLAQANQVASILGDRSEQVDRLLVNAKTLIAAFNERGRAVDALLGNISAF
MLBr_02591|M.leprae_Br4923 LIAQANQVASVLGDRGAQVDRLLVNTKTLIAAFNERGRAVDALLGNVAAF
MAB_4151c|M.abscessus_ATCC_199 LLTHAKSVSGILSERAGQVNKLVTDGNQLFAALDERRLAISELINRIQGL
*:: *:.:: *.:*. *:: *..: : *: *.:** ::. *: .: .
Mflv_0709|M.gilvum_PYR-GCK STQVRGFIDDN-PNLNNVLEQLRVITDVLAARKLDITDTLSSLSKFMVSL
Mvan_0136|M.vanbaalenii_PYR-1 STQVRGFIDDN-PNLNHVLEQLRVISDVLAERKLDLADTLSSLSKFMVSL
MSMEG_0136|M.smegmatis_MC2_155 SRQVEGFVDEN-PNLNHVLEQLRTVSDVLNERKQDLADILTVAGKFITSL
TH_3066|M.thermoresistible__bu STQVRGFIDDN-PNLNTVLEQLRQVSDILVERRYDLVDVLSSLSKFTASL
MMAR_0414|M.marinum_M SAQVQGLINDN-PNLNHVLEQLRTVSDVLIERKEDLAKGLEMVGAFLPSL
MUL_1064|M.ulcerans_Agy99 SAQVQGLINDN-PNLNHVLEQLRTVSDVLIERKEDLAKGLEMVGAFLPSL
MAV_5013|M.avium_104 SEQVKGLINDN-PNLNHVLEQLRTVSDILVQRKDDLANGLTEVGKFLPSL
Mb0177|M.bovis_AF2122/97 SAQVQNLINDN-PNLNHVLEQLRILTDLLVDRKEDLAETLTILGRFSASF
Rv0171|M.tuberculosis_H37Rv SAQVQNLINDN-PNLNHVLEQLRILTDLLVDRKEDLAETLTILGRFSASF
MLBr_02591|M.leprae_Br4923 AAQVQRLINDN-PNLNHVLEQLHQLSGILVQHKDDLANTLIQVAAFLPSL
MAB_4151c|M.abscessus_ATCC_199 SQQISGFVADNRQEFRPTMEKLSVVLDDLLDRKAHISEALKRLPGYGTAL
: *: :: :* ::. .:*:* : . * :: .: . * : ::
Mflv_0709|M.gilvum_PYR-GCK GEALASGPYFKVMLANLAPYWILQPFVDAAFKKRGINPEEFWRNAGLPAF
Mvan_0136|M.vanbaalenii_PYR-1 GEALASGPYFKVMLANLAPYWILQPFVDAAFKKRGINPEEFWRNAGLPAF
MSMEG_0136|M.smegmatis_MC2_155 AEALASGPYFKVMLVNLIPPTILQPFVDAAFKKRGIDPEEFWRNAGLPAF
TH_3066|M.thermoresistible__bu AEGLASGPYFKVMLVNLVPYQILQPFVDAAFKERGIDPENFWRGAGLPEF
MMAR_0414|M.marinum_M NESIASGPFFKVVIHNLVPGQILQPFIDAAFKKRGLSPEDFWRSAGLPEY
MUL_1064|M.ulcerans_Agy99 NESIASGPFFKVVIHNLVPGQILQPFIDAAFKKRGLSPEDFWRSAGLPEY
MAV_5013|M.avium_104 NEAIASGPFFKVVLHNLALYQISQPWVDAAFKKRGIDPEDFWRSAGLPAY
Mb0177|M.bovis_AF2122/97 GETFASGPYFKVLLANLVPGQILQPFVDAAFKKRGISPEDFWRSAGLPAY
Rv0171|M.tuberculosis_H37Rv GETFASGPYFKVLLANLVPGQILQPFVDAAFKKRGISPEDFWRSAGLPAY
MLBr_02591|M.leprae_Br4923 NEAIGSGPFFKVVLHNLAPYQILQPWVDAAFKKRGIDPENFWRSAGLPEF
MAB_4151c|M.abscessus_ATCC_199 GEVVSSGPGFHVNVYGFPPATLTAVAFDTYFQ------------------
* ..*** *:* : .: : .*: *:
Mflv_0709|M.gilvum_PYR-GCK RFPDPNGVGFENGAPPPAPTPIEGTPENPGPAVPPGSPCSYTPPGNG---
Mvan_0136|M.vanbaalenii_PYR-1 RFPDPNGVGFENGAPPPAPTPIEGTPENPGPAVPPGSPCSYTPPADG---
MSMEG_0136|M.smegmatis_MC2_155 RFPDPNGERHENGAPPAAPTPLEGTPEHPGPAVPPGSPCSYTPPADG---
TH_3066|M.thermoresistible__bu QWPDPNAERHANGAPPAAPRPIEGTPEHPGPAVPPGHPCSYTPAANA---
MMAR_0414|M.marinum_M RFPDPNGTRFPNGAPPPGPRVLEGTPDHPLPAVPPGSPCSYTPAADA---
MUL_1064|M.ulcerans_Agy99 RFPDPKGTRFPNGAPPPGPRVLEGTPDHPLPAVPPGSPCSYTPAADA---
MAV_5013|M.avium_104 RFPDPNGTRFPNGAPPPAPPVLEGTPDHPGPAVPPGSPCSYTPAADG---
Mb0177|M.bovis_AF2122/97 RWPDPNGTRFPNGAPPPPPPVLEGTPEHPGPAVPPGSPCSYTPPADG---
Rv0171|M.tuberculosis_H37Rv RWPDPNGTRFPNGAPPPPPPVLEGTPEHPGPAVPPGSPCSYTPPADG---
MLBr_02591|M.leprae_Br4923 RFPDPNGTRFPNGAPPPAPPVLEGTPENPGPAVPPGSPCSYTPAHPGGNN
MAB_4151c|M.abscessus_ATCC_199 ------------------------------PGKLPASLSDYLNG------
*. *. ..*
Mflv_0709|M.gilvum_PYR-GCK -------LPSPGNPLPCASLTQGPYGPVPGG--YPPPN-VVTSEPNVHGP
Mvan_0136|M.vanbaalenii_PYR-1 -------LPRPGNPLPCAGLTQGPYGPVPGG--FPPPN-VATSAPNAHGP
MSMEG_0136|M.smegmatis_MC2_155 -------IPSPGNPLPCAHLSQGPYGPVPGG--YPPPN-VATSAPNPDGI
TH_3066|M.thermoresistible__bu -------APRPGDPLPCADAGVGPFGDNPYGPNYAPPN-VVG-EPHNPDA
MMAR_0414|M.marinum_M -------LPRPDNPLPCAGVLEGPFGGFGFP---APLDGVESSPPNPNGL
MUL_1064|M.ulcerans_Agy99 -------LPRPDNPLPCAGVLEGPFGGFGFP---APLDGVESSPPNPNGL
MAV_5013|M.avium_104 -------LPRPDNPLPCAGAVTGPFGGPGFP---APVD-VMTSPPNPAGL
Mb0177|M.bovis_AF2122/97 -------LPRPWDPLPCANLTQGPFGGPDFP---APLD-VATSPPNPDGP
Rv0171|M.tuberculosis_H37Rv -------LPRPWDPLPCANLTQGPFGGPDFP---APLD-VATSPPNPDGP
MLBr_02591|M.leprae_Br4923 TIGDEMGLPRPWNPLPCAGATAGPYGGPAFP---APID-VQTSPPNPDGL
MAB_4151c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0709|M.gilvum_PYR-GCK QPSPGVPAAAIPGQLSPEVPGAR-PPLPPAPPGARTVPLAPQPAPADFTP
Mvan_0136|M.vanbaalenii_PYR-1 QPSPGVPAAAIPGQLSPAVPGAR-PPLPPAPPGARTVPLEPQPSPPDFTP
MSMEG_0136|M.smegmatis_MC2_155 AHSPGVPSAAIPGQMPPEQPGAP-VEIAPGPPGARTVPVSPIPGAPDFTP
TH_3066|M.thermoresistible__bu QPSPGLPAAAVPGQVSPNVPGAPGPGVAPGPPGX----------------
MMAR_0414|M.marinum_M PPTPGIPIAGRPGEPAPDAPGTPVPLPPDAPPGARTEPVGPA-GPTPPPS
MUL_1064|M.ulcerans_Agy99 PPTPGIPIPGRPGEPAPDAPGTPVPLPPDAPPGARTEPVGPA-GPTPPPS
MAV_5013|M.avium_104 PPTPGIPIAGRPGDAPPDVPGTPVPLPTQAPPGARTENLAPA-GPVPPPS
Mb0177|M.bovis_AF2122/97 PPAPGLPIAGRPGEVPPNVPGTPVPIPQEAPPGARTLPLGPAPGPAPPP-
Rv0171|M.tuberculosis_H37Rv PPAPGLPIAGRPGEVPPNVPGTPVPIPQEAPPGARTLPLGPAPGPAPPP-
MLBr_02591|M.leprae_Br4923 PLLPGIAIAGRPGDPAPNVPGTPVPLPPNAPPGARTEQLAPA-APAQ---
MAB_4151c|M.abscessus_ATCC_199 ----------LIGERQIMRPKSP---------------------------
*: * :
Mflv_0709|M.gilvum_PYR-GCK GIAPLPPALTGPPPPPGPGP-SPGPAGQPLLPGNPPFLPPGSQG-
Mvan_0136|M.vanbaalenii_PYR-1 GIAPLPPALTGPPPPAGPGP-APAPAGQPLLPGNPPFLPPGSQG-
MSMEG_0136|M.smegmatis_MC2_155 GIAPPPPAITGPPPPPGPGP-QLAPVGEAPLPGNPPFLPPGSQSR
TH_3066|M.thermoresistible__bu -------XXARRPARSSPGR-SPAP--------------------
MMAR_0414|M.marinum_M TFAP--GLPPGPPAPPGPGPQLPDPYITPGGTGGSGATGG-SQN-
MUL_1064|M.ulcerans_Agy99 TFAP--GLPPGPPAPPGPGPQLPDPYITPGGTGGSGATGG-SQN-
MAV_5013|M.avium_104 TFAP--GLPPGPPAPPGPGNQLPAPFINPGGTGGSGAAGGGSQN-
Mb0177|M.bovis_AF2122/97 -------AAPGPPAPPGPGPQLPAPFINPGGTGGSGVTGG-SEN-
Rv0171|M.tuberculosis_H37Rv -------AAPGPPAPPGPGPQLPAPFINPGGTGGSGVTGG-SEN-
MLBr_02591|M.leprae_Br4923 ----------DSSAPHEPREQLSAPLINPGGTGGSGGAGG-SRN-
MAB_4151c|M.abscessus_ATCC_199 ---------------------------------------------