For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTPQTGSHTSVRASGRPAPVLERNYDRPAALDNPRAPRRGSGMPNFEKYAWIFMRLSGVVLIFLALGHL FIMLMWDNGVYRLDFNFVAQRWASPFWQTWDLLMLWLAQLHGGNGMRTIIADYTRKDSTRFWLNCLLALS IIFTVVLGTYVLLTFNPNIG
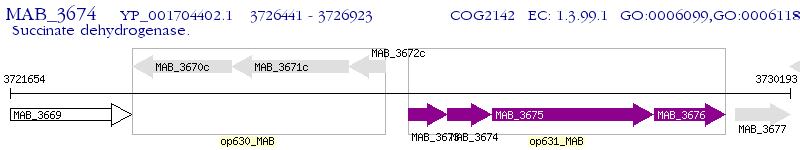
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3674 | - | - | 100% (160) | succinate dehydrogenase, hydrophobic membrane anchor protein SdhD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3346 | sdhD | 1e-70 | 80.85% (141) | succinate dehydrogenase hydrophobic membrane anchor subunit |
| M. gilvum PYR-GCK | Mflv_4845 | - | 1e-72 | 81.94% (144) | putative succinate dehydrogenase |
| M. tuberculosis H37Rv | Rv3317 | sdhD | 2e-70 | 80.14% (141) | succinate dehydrogenase hydrophobic membrane anchor subunit |
| M. leprae Br4923 | MLBr_00698 | sdhD | 1e-65 | 74.31% (144) | putative succinate dehydrogenase hydrophobic membrane anchor |
| M. marinum M | MMAR_1202 | sdhD | 5e-69 | 78.47% (144) | succinate dehydrogenase (hydrophobic membrane anchor |
| M. avium 104 | MAV_4298 | - | 5e-67 | 77.78% (144) | succinate dehydrogenase hydrophobic membrane anchor protein |
| M. smegmatis MC2 155 | MSMEG_1671 | - | 2e-63 | 71.53% (144) | succinate dehydrogenase hydrophobic membrane anchor protein |
| M. thermoresistible (build 8) | TH_2181 | sdhD | 1e-70 | 84.17% (139) | PROBABLE SUCCINATE DEHYDROGENASE (HYDROPHOBIC MEMBRANE |
| M. ulcerans Agy99 | MUL_1368 | sdhD | 9e-67 | 78.42% (139) | succinate dehydrogenase (hydrophobic membrane anchor |
| M. vanbaalenii PYR-1 | Mvan_1588 | - | 3e-72 | 81.25% (144) | putative succinate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4845|M.gilvum_PYR-GCK MTQASRPASGRAENAYDHISDRGGPAPVMQRSFDRPAGLDNPRAPRRSGG
Mvan_1588|M.vanbaalenii_PYR-1 MSSGKK----MAENPYDHISDRGGPAPVMQRSYDRPAGLDNPRSPRRAGG
TH_2181|M.thermoresistible__bu ---------------------------VLETSHDRPAALDNPRAPRRRGG
MSMEG_1671|M.smegmatis_MC2_155 MS---------APGAGE--SRLGRPAPVMEREHDRPAALDHPRAPRKPRG
Mb3346|M.bovis_AF2122/97 MS-----------------------APVRQRSHDRPASLDNPRSPRRRAG
Rv3317|M.tuberculosis_H37Rv MS-----------------------APVRQRSHDRPASLDNPRSPRRRAG
MLBr_00698|M.leprae_Br4923 MSNSDL----QLRPQVQPTCQKGQLAPVRQRSYDRPASLDNPRSPRRRAG
MAV_4298|M.avium_104 MSTPDL----QLGP--------GQIAPVKQRFYDRPASLDNPRSPRRRAG
MMAR_1202|M.marinum_M MSAPDH----QLGR--------GEHAPVLQRSHDRPASLDNPRSPRRRAG
MUL_1368|M.ulcerans_Agy99 ---------------------------MLQRSHDRPASLDNPRSPRRRAS
MAB_3674|M.abscessus_ATCC_1997 MTTP-------QTGSHTSVRASGRPAPVLERNYDRPAALDNPRAPRRGSG
: : .****.**:**:**: .
Mflv_4845|M.gilvum_PYR-GCK MPNFEKYTWLFMRFSGIVLVFLALGHLFIMLMWENGVYRIDFNYVAQRWS
Mvan_1588|M.vanbaalenii_PYR-1 MPNFEKYAWLFMRFSGIVLVFLALGHLFIMLMWENGVYRIDFNYVAERWS
TH_2181|M.thermoresistible__bu MPNFEKYAWLFMRFSGIVLVFLALGHLFIMLMWENGVYRIDFNYVAERWQ
MSMEG_1671|M.smegmatis_MC2_155 IPYFEKYAWLFMRFSGIALVFLALGHLFIMLMWQDGVYRIDFNYVAERWA
Mb3346|M.bovis_AF2122/97 MPNFEKFAWLFMRFSGVVLVFLAIGHLFIMLMWDNGVYRLDFNFVAQRWA
Rv3317|M.tuberculosis_H37Rv MPNFEKFAWLFMRFSGVVLVFLAIGHVFIMLMWDNGVYRLDFNFVAQRWA
MLBr_00698|M.leprae_Br4923 IPNFEKFAWLFMRFSGVALVVLAFGHLFIMLMWDNGVYRIDFNFVAQRWS
MAV_4298|M.avium_104 IPNFEKFAWLFMRFSGVALFVLAIGHLFIMLMWDDGVYRIDFNYVAQRWA
MMAR_1202|M.marinum_M MPNFEKYAWLFMRFSGVVLVFLALGHLFIGLMWENGVYRIDFNYVAQRWS
MUL_1368|M.ulcerans_Agy99 MPNFEKYAWLFMRFSGVVLVFLALGHLFIGLMWENGVYRIDFNYVAQRWS
MAB_3674|M.abscessus_ATCC_1997 MPNFEKYAWIFMRLSGVVLIFLALGHLFIMLMWDNGVYRLDFNFVAQRWA
:* ***::*:***:**:.*..**:**:** ***::****:***:**:**
Mflv_4845|M.gilvum_PYR-GCK SPFWQTWDLLLLWLAQLHGGNGMRTIIADYTRKDSTKFWLNTLLALSMAF
Mvan_1588|M.vanbaalenii_PYR-1 SPFWQTWDLLLLWLAQLHGGNGMRTIIADYSRKDSTKFWLNTLLALSMAF
TH_2181|M.thermoresistible__bu SPFWQIWDLLLLWLAQLHGGNGMRTIIADYARKDSTRFWLNALLALSIVF
MSMEG_1671|M.smegmatis_MC2_155 SPFWQIWDMALLWLAMIHGANGMRTIIGDYARKNVTKFWLNSLLLLATGF
Mb3346|M.bovis_AF2122/97 SPFWQTWDLLLLWLAQLHGGNGLRTIIDDYSRKDTTRFWLNSLLVLSMLF
Rv3317|M.tuberculosis_H37Rv SPFWQTWDLLLLWLAQLHGGNGLRTIIDDYSRKDTTRFWLNSLLVLSMLF
MLBr_00698|M.leprae_Br4923 SPFWRFWDLALLWLAQVHGGNGLRNIIDDYSRKDHTRFWLNSLLLLSMGF
MAV_4298|M.avium_104 SPFWQFWDLALLWLAQLHGGNGLRTIIDDYSRKDSTRFWLNSLLLLSMGF
MMAR_1202|M.marinum_M SPFWQTWDLLLLWLAQLHGGNGLRVIIDDYSRKDSTRFWLNSLLVVSIVF
MUL_1368|M.ulcerans_Agy99 SPFWQTWDLLLLWLAQLHGGNGLRVIIDDYSRKDSTRFWLNSLLVVSIVF
MAB_3674|M.abscessus_ATCC_1997 SPFWQTWDLLMLWLAQLHGGNGMRTIIADYTRKDSTRFWLNCLLALSIIF
****: **: :**** :**.**:* ** **:**: *:**** ** :: *
Mflv_4845|M.gilvum_PYR-GCK TLVLGTYVLLTFDASIS-
Mvan_1588|M.vanbaalenii_PYR-1 TLVLGTYVLLTFDATIS-
TH_2181|M.thermoresistible__bu TLVLGTYVLLTFDANIS-
MSMEG_1671|M.smegmatis_MC2_155 TLVLGSYVLVTFDANIS-
Mb3346|M.bovis_AF2122/97 TLMLGTYVIVTFDPNIS-
Rv3317|M.tuberculosis_H37Rv TLMLGTYVIVTFDPNIS-
MLBr_00698|M.leprae_Br4923 TLVLGSYVLLSFDPNIS-
MAV_4298|M.avium_104 TLVLGTYVLLTFDPNISG
MMAR_1202|M.marinum_M TLALGSYVLLTFDANIS-
MUL_1368|M.ulcerans_Agy99 TLALGSYVLLTFDANIS-
MAB_3674|M.abscessus_ATCC_1997 TVVLGTYVLLTFNPNIG-
*: **:**:::*:..*.