For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTNTLTPRDEKKAAKSKSLAKHETGVGLQKHKRGAIDILIAVLTPIAGSELLDKYNLRGAFNKGIFESTK GLFTTLGVANRTFKKVTGSKGGPKRLDKANADYFDLNPEDEQKMIADTVKEFAVEVIRPAAYESDKNKTY PADLLGKVAELGVTAINIPEDFDGIASQRSTVTNSLVAEALSYGDMGLALPILAPSGVASALTNWGSADQ QATYLKEFAGENVPQASVVIAEPQALFDPFSLKTTATRTPSGFRLNGTKSLVPAAAQAELFIVAANYNGR PTLFIVESGTEGITVEEDPSMGIRGAALGRLNLNNVAVPAANILGEEEGSDTALANYSEAVALARLGWAS LAVGTGQAVLDYVIPYVKQREAFGEPIARRQAVAFMCANIAIELDGLRLVTLRGASRAEQGLPFVREAAL ARKLATDKGMQIGLDGVQLLGGHGFTKEHPVERWYRNLRAIGVAEGVVVL
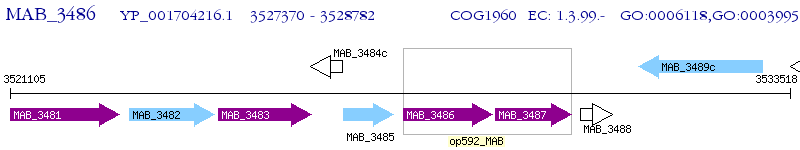
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3486 | - | - | 100% (470) | acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1188c | - | 6e-39 | 30.25% (367) | acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4538c | - | 2e-36 | 28.73% (369) | putative acyl-CoA dehydrogenase FadE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3163 | fadE24 | 0.0 | 69.43% (471) | acyl-CoA dehydrogenase FADE24 |
| M. gilvum PYR-GCK | Mflv_4455 | - | 1e-180 | 68.63% (475) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3139 | fadE24 | 0.0 | 69.43% (471) | acyl-CoA dehydrogenase FADE24 |
| M. leprae Br4923 | MLBr_00661 | fadE24 | 1e-174 | 67.34% (447) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_1510 | fadE24 | 0.0 | 70.70% (471) | acyl-CoA dehydrogenase FadE24 |
| M. avium 104 | MAV_4018 | - | 0.0 | 70.54% (448) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2081 | - | 0.0 | 70.34% (445) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3828 | - | 0.0 | 69.36% (470) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_2431 | fadE24 | 0.0 | 69.21% (471) | acyl-CoA dehydrogenase FadE24 |
| M. vanbaalenii PYR-1 | Mvan_1907 | - | 1e-177 | 66.38% (473) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3163|M.bovis_AF2122/97 MTNTTSAANAAKPSGARTDRRGRTTGVGLAPHKRTGIDVALALLTPIVGQ
Rv3139|M.tuberculosis_H37Rv MTNTTSAANAAKPSGARTDRRGRTTGVGLAPHKRTGIDVALALLTPIVGQ
MLBr_00661|M.leprae_Br4923 MTDT---APSAKPSSVRLKRGGLRSGVGMQPHKRTGFDVAIAMLTPLVGQ
MAV_4018|M.avium_104 MTDS---GSTKTP--ARVKRRDRQSGVGLQPHKRTGIDIAIALLTPLVGQ
MMAR_1510|M.marinum_M MTETLP-APSAKRSRSRDDR---QTSLGMQPHKRTGIDVALAFLTPIIGQ
MUL_2431|M.ulcerans_Agy99 MTETLP-APSAKRSRSRDDR---QTSLGMQPHKRTGIDVALAFLTPIIGQ
Mflv_4455|M.gilvum_PYR-GCK MTNTLP-----------PKRGDRESAVGLQKHRRTAIDVGMALLTPLMGQ
Mvan_1907|M.vanbaalenii_PYR-1 MTNTLP-----------SKRGGRESAVGLQTHKRSVTDIGLALITPIVGQ
MSMEG_2081|M.smegmatis_MC2_155 --------------------------MGLQKHKRTATDIGLALITPIVGQ
TH_3828|M.thermoresistible__bu MTNTLP---STNGSTPRRKRTGPESAVGLQRHKRTATDIGLALITPLVGQ
MAB_3486|M.abscessus_ATCC_1997 MTNTLT-PRDEKKAAKSKSLAKHETGVGLQKHKRGAIDILIAVLTPIAGS
:*: *:* *: :*.:**: *.
Mb3163|M.bovis_AF2122/97 EFLDKYRLRDPLNRSLRYGVKTMFATAGAATRQFQRVQGLRGG-PTRLKS
Rv3139|M.tuberculosis_H37Rv EFLDKYRLRDPLNRSLRYGVKTMFATAGAATRQFQRVQGLRGG-PTRLKS
MLBr_00661|M.leprae_Br4923 DFLDKYHLRYPLNRALRYGVKATFSTAAATSRQFKRVQNLRGG-ATRLQP
MAV_4018|M.avium_104 EFLDKYHLRDPLNRGLRYGTKTIFSTGAATSRQFKRVQNLRGG-PTRLKS
MMAR_1510|M.marinum_M EFLDKYNLRDPLNRGLRYGVKTMFSTSGKANRQFQRVQNLRGGGPVRLKS
MUL_2431|M.ulcerans_Agy99 EFLDKYTLRDPLNRGLRYGVKTMFSTSGKANRQFQRVQNLRGGGPVRLKS
Mflv_4455|M.gilvum_PYR-GCK EFLDKYNLRDPLNRGLKYGVKHGFSAAGAATRQFKRVQGIGKP-ATRLTE
Mvan_1907|M.vanbaalenii_PYR-1 EFLDRYNLREPLNRGIRYGVKHAFSAAGASTRQFKRIQGLGKP-ATRLEA
MSMEG_2081|M.smegmatis_MC2_155 EWLDRYGLRDPLNRGLKYGVKQVFSTAGATTRQFKRIQGIGKA-PTRLKA
TH_3828|M.thermoresistible__bu EFLDKYNLRDPLNRALRYGVKTAFSTAGAATRQFKRVQGLRGE-PTRLKP
MAB_3486|M.abscessus_ATCC_1997 ELLDKYNLRGAFNKGIFESTKGLFTTLGVANRTFKKVTGSKGG-PKRLDK
: **:* ** .:*:.: ..* *:: . :.* *::: . . **
Mb3163|M.bovis_AF2122/97 S---GRDYFDLTPDDDQKLIIETVDEFAEEVLRPAAHDADDAATYPSDLT
Rv3139|M.tuberculosis_H37Rv S---GRDYFDLTPDDDQKLIIETVDEFAEEVLRPAAHDADDAATYPSDLT
MLBr_00661|M.leprae_Br4923 S---GRNYFDLTPDDNQKLIVETIDEFSAEILRPAAHDADEAAAYPPDLI
MAV_4018|M.avium_104 S---GKDYFDLTPDDEQKMIVETLDEFAAEVLRPAAHDADEAATYPRDLI
MMAR_1510|M.marinum_M S---GKDYFDLTPDDDQKMIVETVQEFAAEILRPAAHDADEAISYPADLI
MUL_2431|M.ulcerans_Agy99 S---GKDYFDLTPDDDQKMIVETVQEFAAEILRPAAHDADEAISYPADLI
Mflv_4455|M.gilvum_PYR-GCK NRPKGSDYFDLTPDDDQKMIVQTVEEFAEEILRPAAHDADDAATYPPDLI
Mvan_1907|M.vanbaalenii_PYR-1 HRPQGSDYFDLTPDEDQKMIVETVEEFAEEILRPAAYDADDAASYPPDLI
MSMEG_2081|M.smegmatis_MC2_155 S---GADLFDLTPDDDQKMIVETVNEFAAEILRPAAEEADEAATYPPDLI
TH_3828|M.thermoresistible__bu S---GSDYFDLTPDDEQQMIVDTVSEFAKEVLRPAAREADDAARFPADLM
MAB_3486|M.abscessus_ATCC_1997 A---NADYFDLNPEDEQKMIADTVKEFAVEVIRPAAYESDKNKTYPADLL
. : ***.*:::*::* :*:.**: *::**** ::*. :* **
Mb3163|M.bovis_AF2122/97 AKAAELGITAINIPEDFDGIAEHRSSVTNVLVAEALAYGDMGLALPILAP
Rv3139|M.tuberculosis_H37Rv AKAAELGITAINIPEDFDGIAEHRSSVTNVLVAEALAYGDMGLALPILAP
MLBr_00661|M.leprae_Br4923 AKAAKLGITTINIPEYFDGIAERRSSVTNVLVAEALAYGDMGLTLPILAP
MAV_4018|M.avium_104 AKAAELGITAINIPEDFDGIAAHRSSVTNVLVAEALAYGDMGLALPLLAP
MMAR_1510|M.marinum_M AKAAELGITAINIPEDFDGIAEQRSTVTNALVAEALAYGDMGLALPILAP
MUL_2431|M.ulcerans_Agy99 AKAAELGITAINIPEDFDGIAEQRSTVTNALVAEALAYGDMGLALSILAP
Mflv_4455|M.gilvum_PYR-GCK AKAAELGITAINIPEDFDGIAEHRSTVTNALVAEALAYGDMGLALPILAP
Mvan_1907|M.vanbaalenii_PYR-1 AKAAELGITAINVPEDFDGIAEHRTTVTGALVAEALAYGDMGLALPILAP
MSMEG_2081|M.smegmatis_MC2_155 AKAAELGITAVNVPEDFEGIAEHRSTVTNALVAEALAYGDMGLALPILAP
TH_3828|M.thermoresistible__bu GKAAELGITALNIPENFDGIAAHRSTVTSALVAEALAYGDMGLALPILAP
MAB_3486|M.abscessus_ATCC_1997 GKVAELGVTAINIPEDFDGIASQRSTVTNSLVAEALSYGDMGLALPILAP
.*.*:**:*::*:** *:*** :*::**. ******:******:*.:***
Mb3163|M.bovis_AF2122/97 GGVASALTHWGSADQQATYLKEFAGENVPQACVAITEPQPLFDPTRLKTT
Rv3139|M.tuberculosis_H37Rv GGVASALTHWGSADQQATYLKEFAGENVPQACVAITEPQPLFDPTRLKTT
MLBr_00661|M.leprae_Br4923 GGVAATLTYWGSADQQATYLKEFTGENVPQACVAIAEPQPLFDPTRLKTI
MAV_4018|M.avium_104 GGVASALTHWGSAEQQATYLPEFAGENVPQACVAIAEPQPLFDPTRLKTS
MMAR_1510|M.marinum_M GGVASALTHWGSADQQATYLSEFAGENVPQACVAIAEPQPLFDPTRLKTT
MUL_2431|M.ulcerans_Agy99 GGVASALTHWGSADQQATYLSEFAGDNVPQACVAITEPQPLFDPTRLKTT
Mflv_4455|M.gilvum_PYR-GCK GGVASALTHWGSADQQATYLREFAGESVPQACLAIAEPHPLFDPTALRTT
Mvan_1907|M.vanbaalenii_PYR-1 GGVASALTHWGSADQQATYLKEFSGDNVPQACVAIAEPHPLFDPTALKTT
MSMEG_2081|M.smegmatis_MC2_155 GGVASALTHWGSADQQATYLKEYAGDNVPQSSVVIAEPHALFDPTELKTT
TH_3828|M.thermoresistible__bu GGVAAALTHWGSADQQATYLPEYAGENVPQSAVVIAEPQPLFDPTMLKTT
MAB_3486|M.abscessus_ATCC_1997 SGVASALTNWGSADQQATYLKEFAGENVPQASVVIAEPQALFDPFSLKTT
.***::** ****:****** *::*:.***:.:.*:**:.**** *:*
Mb3163|M.bovis_AF2122/97 AVRTPSGYRLDGVKSLIPAAADAELFIVGAQLGGKPALFIVESAASGLTV
Rv3139|M.tuberculosis_H37Rv AVRTPSGYRLDGVKSLIPAAADAELFIVGAQLGGKPALFIVESAASGLTV
MLBr_00661|M.leprae_Br4923 AVRTPSGYRLDGVKSLVPAAASAELFIIGAQLNGKPALFIVESSTKGLTV
MAV_4018|M.avium_104 AVRTPSGYRLDGVKSLVPAAADAELFIVGAQLDGKPALFIVESSTKGLTV
MMAR_1510|M.marinum_M AVRTPSGYRLDGVKSLVPAAADAELFIVAAQLDGKPALFIVESSAAGLTV
MUL_2431|M.ulcerans_Agy99 AVRTPSGYRLDGVKSLVPAAADAELFIVAAQLDGKPALFIVESSAAGLIV
Mflv_4455|M.gilvum_PYR-GCK AVRTPSGYRLDGVKSLVPAAADAELFIIAAALGGKPALFLVEASTPGLTV
Mvan_1907|M.vanbaalenii_PYR-1 AVRTPSGYRLDGVKSLVPAAADAEVFIIAAQLNGKPALFIVEASTKGLTV
MSMEG_2081|M.smegmatis_MC2_155 AVRTPSGYRLSGVKSLVPAAADAELFIVAAQFNGKPALFIVEASSKGLSV
TH_3828|M.thermoresistible__bu AVRTPSGYRLHGVKSLVPAAADAELFIVGAQLSGRPTLFIVESDTAGLSV
MAB_3486|M.abscessus_ATCC_1997 ATRTPSGFRLNGTKSLVPAAAQAELFIVAANYNGRPTLFIVESGTEGITV
*.*****:** *.***:****.**:**:.* .*:*:**:**: : *: *
Mb3163|M.bovis_AF2122/97 KADPSMGIRGAALGQVELCGVSVPLNARLGEDEASDND----YSEALALA
Rv3139|M.tuberculosis_H37Rv KADPSMGIRGAALGQVELCGVSVPLNARLGEDEASDND----YSEALALA
MLBr_00661|M.leprae_Br4923 KADPSMGIRAAALGQVELCGVSAPLDARLGEDDATDND----YSEAIALA
MAV_4018|M.avium_104 KADPSMGIRAAELGHVELSGVTVPLGARLGEDEATDDD----YSEAIALS
MMAR_1510|M.marinum_M KADPSMGIRAAALGQIELSKVSVPLSARLGEDQATDAD----YSEAIALA
MUL_2431|M.ulcerans_Agy99 KADPSMGIRAAALGQIELSKVSVPLRARLGEDQATDAD----YSEAIALA
Mflv_4455|M.gilvum_PYR-GCK TADPSMGIRAAALGRVELAKVTVPLSARLGEEDASAADTAQIYSDAIALS
Mvan_1907|M.vanbaalenii_PYR-1 TPDPSMGIRAAALGRVELDKVTVPLSARLGEEDATDAD----YSEAIALS
MSMEG_2081|M.smegmatis_MC2_155 KADRSMGLRAAALGQIELDNVTVPLSARLGEDGATDQD----YSEAIALA
TH_3828|M.thermoresistible__bu KADPSMGLRPAALGRLQLDNVTVPLSARLGEDVSEQEHDQN-YSEAVALA
MAB_3486|M.abscessus_ATCC_1997 EEDPSMGIRGAALGRLNLNNVAVPAANILGEEEGSDTALAN-YSEAVALA
* ***:* * **:::* *:.* ***: . **:*:**:
Mb3163|M.bovis_AF2122/97 RLGWAALAVGTSHAVLDYVVPYVKQRQAFGEPIAHRQAVAFMCANIAIEL
Rv3139|M.tuberculosis_H37Rv RLGWAALAVGTSHAVLDYVVPYVKQRQAFGEPIAHRQAVAFMCANIAIEL
MLBr_00661|M.leprae_Br4923 RLGWAALAVGTSHAVLDYVMPYVKEREAFGEPIARRQAVAFMCANIAIEL
MAV_4018|M.avium_104 RLGWAALTVGTSHAVLDYVVPYVKEREAFGEPIARRQAVAFMCANIAIEL
MMAR_1510|M.marinum_M RLGWAALAVGTSHAVLDYVIPYVKEREAFGEPIARRQAVAFMCANIAIEL
MUL_2431|M.ulcerans_Agy99 RLGWAALAVGTSHAVLDYVIPYVKERKAFGEPIARRQAVAFMCANIAIEL
Mflv_4455|M.gilvum_PYR-GCK RLGWAALAVGTSHAVLDYVIPYVKEREAFGEPIARRQSVAFMCANIAIEL
Mvan_1907|M.vanbaalenii_PYR-1 RLGWAALAVGTSHAVLDYVIPYVKERTAFGEPIAHRQSVAFMCANIAIEL
MSMEG_2081|M.smegmatis_MC2_155 RLGWAALAVGTSHAVLDYVVPYVKERHAFGEPIAHRQSVAFMCANIAIEL
TH_3828|M.thermoresistible__bu RLGWAALAVGTSHAVLDYVVPYVKERHAFGEPIAHRQAVAFMTADIAIEL
MAB_3486|M.abscessus_ATCC_1997 RLGWASLAVGTGQAVLDYVIPYVKQREAFGEPIARRQAVAFMCANIAIEL
*****:*:***.:******:****:* *******:**:**** *:*****
Mb3163|M.bovis_AF2122/97 DGLRLITWRGASRAEQGLPFAREAALAKRLGSDKGMQIGLDGVQLLGGHG
Rv3139|M.tuberculosis_H37Rv DGLRLITWRGASRAEQGLPFAREAALAKRLGSDKGMQIGLDGVQLLGGHG
MLBr_00661|M.leprae_Br4923 DGLRLITWRGASRAEQGLSFAREAALAKRLGSDKAMQIGLDGVQLLGGHG
MAV_4018|M.avium_104 DGLRLITWRGAARAEQGLPFAREAALAKRLGADKGMQIGSDGVQLLGGHG
MMAR_1510|M.marinum_M DGLRLITWRGASRAEQGLPFAREAALAKRLGSDKGMQIGLDGVQLLGGHG
MUL_2431|M.ulcerans_Agy99 DGLRLITWRGASRAEQGLSFAREAALAKRLGSDKGMQIGLDGVQLLGGHG
Mflv_4455|M.gilvum_PYR-GCK DGLRLITWRGASRAEQGLSFAREAALAKKLGADKGMQIGLDGVQLLGGHG
Mvan_1907|M.vanbaalenii_PYR-1 DGLRLITWRGAARADQGMSFAREAALAKKLGSDKAMQIGLDGVQLLGGHG
MSMEG_2081|M.smegmatis_MC2_155 DGLRLITWRGAARAEQGLPFAREAALARKLGADKGMQIGLDGVQLLGGHG
TH_3828|M.thermoresistible__bu DGLRLITWRGASRAEQGMPFIREAALAKRFATDKGMRIGLDGVQLLGGHG
MAB_3486|M.abscessus_ATCC_1997 DGLRLVTLRGASRAEQGLPFVREAALARKLATDKGMQIGLDGVQLLGGHG
*****:* ***:**:**:.* ******:::.:**.*:** **********
Mb3163|M.bovis_AF2122/97 YTKEHPVERWYRDLRAIGVAEGVVVI
Rv3139|M.tuberculosis_H37Rv YTKEHPVERWYRDLRAIGVAEGVVVI
MLBr_00661|M.leprae_Br4923 FIKEHPVERWYRDLRAIGVAEGVVVI
MAV_4018|M.avium_104 YTNEHPVERWYRDLRAIGVAEGVVVI
MMAR_1510|M.marinum_M YTKEHPVERWYRDLRAIGVAEGVVVI
MUL_2431|M.ulcerans_Agy99 YTKEHPVERWYRDLRAIGVAEGVVVI
Mflv_4455|M.gilvum_PYR-GCK YTKEHPVERWYRDLRAIGVAEGVVVL
Mvan_1907|M.vanbaalenii_PYR-1 YTKEHPVERWYRDLRALGVAEGVVVL
MSMEG_2081|M.smegmatis_MC2_155 YTKEHPVERWYRDLRAIGIAEGVVVI
TH_3828|M.thermoresistible__bu YTKEHPVERWYRDLRAIGVAEGVVVL
MAB_3486|M.abscessus_ATCC_1997 FTKEHPVERWYRNLRAIGVAEGVVVL
: :*********:***:*:******: