For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKNSVALGMCFSAALLCAATASTGCAAAAPPSGFPDISGLPAVDSRQYFSVKVYRFATPEGVLCESAGWN PYFQWTCVGPRGAGAAQLDLGTYARNNGTPATVSESQSNPDVSRVPVLPANHQVRIAAPVHNNDGVQTIL TCATPAAKTTACLVDDLHGTHGFVLSPERNWAF
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
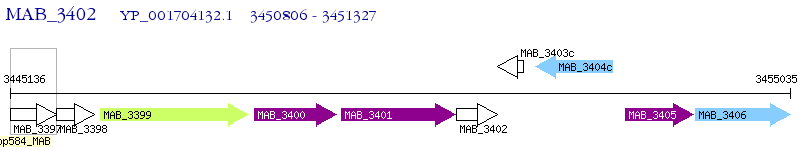
| M. abscessus ATCC 19977 | MAB_3402 | - | - | 100% (173) | hypothetical protein MAB_3402 |
| M. abscessus ATCC 19977 | MAB_0733 | - | 3e-10 | 28.65% (171) | hypothetical protein MAB_0733 |
| M. abscessus ATCC 19977 | MAB_3472 | - | 2e-06 | 25.54% (184) | hypothetical protein MAB_3472 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3413 | - | 2e-06 | 28.65% (178) | hypothetical protein Mflv_3413 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0501 | - | 3e-05 | 25.26% (194) | hypothetical protein MAV_0501 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4865 | - | 7e-05 | 29.49% (156) | hypothetical protein Mvan_4865 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3402|M.abscessus_ATCC_1997 MKNSVALGMCFSAALLCAATAS------TGCAAAAPPSGFPDISGLPAVD
Mflv_3413|M.gilvum_PYR-GCK --MTAWVRASVLAAGVAGAVVG------ASPSTAAPPPDFPDLAAYPAVD
MAV_0501|M.avium_104 ----MLSKTFNATAAVIGTAVI------SACTAHAEPPKFPDLDAFQPVD
Mvan_4865|M.vanbaalenii_PYR-1 MRVTIPLIALATAALVAGCSMEKQPAPASVSTETAPPTDTPPADPPPGVD
:* : . : : * *. * **
MAB_3402|M.abscessus_ATCC_1997 ---------------SRQYFSVKVYRFATPEGVLCESAG---WNPYFQWT
Mflv_3413|M.gilvum_PYR-GCK PAAYQ---------VTGGHPSTSGWAFTTAGGLRCQNS----LIPDLGIF
MAV_0501|M.avium_104 PAP-----------YTASGRAGGGAYFVTPDGIQCSVPHPDHPGEHVSVS
Mvan_4865|M.vanbaalenii_PYR-1 FPDLSGYTESFDQFEQTDVPRVQGFSFSTPSGLICSSNAYP-EPQFESVS
* *. *: *.
MAB_3402|M.abscessus_ATCC_1997 CVGPR-GAGAAQLDLGTYARNNGTPATVSESQSN-PDVSRVPVLPANHQV
Mflv_3413|M.gilvum_PYR-GCK CEGP---VPDAPSEVMSVNVSLTTPAGFDRREGE-LETPWSSLLPVGSRF
MAV_0501|M.avium_104 CAGPLPGLPDDAPRGKDGCSAVGSPTSLPTDLG-----PYSFQEGTGCPI
Mvan_4865|M.vanbaalenii_PYR-1 CRGP---IAAQGPGLWEVTTLFGQAATVVAVVDDGLPRDTPPVLPPFHKV
* ** .: . . .
MAB_3402|M.abscessus_ATCC_1997 RIAAPVHNNDGVQTI-LTCATPAAK----TTACLVDDLHGTHGFVLSPER
Mflv_3413|M.gilvum_PYR-GCK AAGNGVVCAAPDEVT-FACWAAKPDSWPADTPDPPDRHYGEHGFVLSPAG
MAV_0501|M.avium_104 LTTPPLNVGQKITTAGITCAVGGDR----LTACIDPVLN--RGFVLEPSG
Mvan_4865|M.vanbaalenii_PYR-1 TASKGAAECGVDDEGTVACRIGQDG------------------FVLTPSS
.:* *** *
MAB_3402|M.abscessus_ATCC_1997 NWAF
Mflv_3413|M.gilvum_PYR-GCK NRGY
MAV_0501|M.avium_104 SWTF
Mvan_4865|M.vanbaalenii_PYR-1 VELF
: