For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAWVRASVLAAGVAGAVVGASPSTAAPPPDFPDLAAYPAVDPAAYQVTGGHPSTSGWAFTTAGGLRCQN SLIPDLGIFCEGPVPDAPSEVMSVNVSLTTPAGFDRREGELETPWSSLLPVGSRFAAGNGVVCAAPDEVT FACWAAKPDSWPADTPDPPDRHYGEHGFVLSPAGNRGY
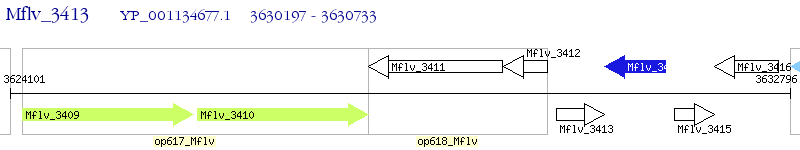
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3413 | - | - | 100% (178) | hypothetical protein Mflv_3413 |
| M. gilvum PYR-GCK | Mflv_5250 | - | 1e-10 | 31.45% (159) | hypothetical protein Mflv_5250 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0733 | - | 4e-49 | 55.36% (168) | hypothetical protein MAB_0733 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0501 | - | 5e-08 | 30.05% (183) | hypothetical protein MAV_0501 |
| M. smegmatis MC2 155 | MSMEG_1136 | - | 2e-07 | 27.12% (177) | hypothetical protein MSMEG_1136 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1273 | - | 2e-09 | 31.74% (167) | hypothetical protein Mvan_1273 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3413|M.gilvum_PYR-GCK MTAWVRASVLAAGVAGAVVGASP-------STAAPPPDFPDLAAYPAVDP
MAB_0733|M.abscessus_ATCC_1997 ----------MAGAVLAAFAVVP-------VANAQPPKFPDVDSYPAVDL
Mvan_1273|M.vanbaalenii_PYR-1 --MRQRLRWLLAGLGAAVIMAAP-------ASAAPPGGDDASGDIDAYPL
MSMEG_1136|M.smegmatis_MC2_155 --MLSRLLVGLASAAGAAVMSVPGVAAADPQADPPPPTVPDVNAYPPVKT
MAV_0501|M.avium_104 --MLSKTFNATAAVIGTAVISAC-------TAHAEPPKFPDLDAFQPVDP
*. :.. : . * .
Mflv_3413|M.gilvum_PYR-GCK AAYQVTGGHPSTSGWAFTTAGGLRCQNSLIPDL----GIFCEGPVPDAPS
MAB_0733|M.abscessus_ATCC_1997 AEYQVIGAHPSMSGWVFSTPGGLRCQSNMIADL----GVSCTGPIVGAAT
Mvan_1273|M.vanbaalenii_PYR-1 AEGDYT--TPDDYGWVFFTTPDGRACGIGPNGG----PVGCDAVAADAPP
MSMEG_1136|M.smegmatis_MC2_155 SEYAVM----DNNWYAFSIGEGITCVLQRTG------SYGCSGPIPAAPN
MAV_0501|M.avium_104 APYTASG--RAGGGAYFVTPDGIQCSVPHPDHPGEHVSVSCAGPLPGLPD
: * . . * . .
Mflv_3413|M.gilvum_PYR-GCK EVMSVNVSLTTPAGFDRREGELETPWSSLLPVGSRFAAGNGVVCAAPDEV
MAB_0733|M.abscessus_ATCC_1997 DMNSVAVSLTKPGLIARYEQSPDDHSYPLLPTGSKIAPGNGVVCAVLADD
Mvan_1273|M.vanbaalenii_PYR-1 GVNQTWVMSWGPAEYRYSDTASFTRGVDMLPVGERVES-LGGACAVTSPD
MSMEG_1136|M.smegmatis_MC2_155 GANLVSGGPGVPG-FASSPAPVFAVVGDAKPLPPNTRLSYQTISCGFDGV
MAV_0501|M.avium_104 DAPRGKDGCSAVGSPTSLPTDLGPYSFQEGTGCPILTTPPLNVGQKITTA
. .
Mflv_3413|M.gilvum_PYR-GCK TFACWAAKPDSWPADTPDPPDRHYGEHGFVLSPAGNRGY
MAB_0733|M.abscessus_ATCC_1997 ALACRAKKPDSWPKDTQDPPDRHYGEHGFVVQPSGSWSY
Mvan_1273|M.vanbaalenii_PYR-1 TVRCET-----------------YGNHGFILSADQGVFW
MSMEG_1136|M.smegmatis_MC2_155 TTACIDGRNQAGFVISPTG-SFVIGQQESDFKPTFQIG-
MAV_0501|M.avium_104 GITCAVGGDRLTACIDP------VLNRGFVLEPSGSWTF
* :: ...