For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RWSTGAFAALGGITAAVATGGTALRSPVVVDRLNDWAARRLTVQQRADPYAAILPTPISGDDFYGDPGDL GLLAPGEVVRADRVTPRLPLRRATMQRIMVRSTDTAGNPVPVTAALIEPERPWRGPGSRPVVVRNQAINS LGLKFTPSYRLTHLWYRDNPPMFPFLSAQNYAVLFPDHEGPRMSYAAGKMAGHAVLDSVRGMLSERPDLA ESPIVMHGYSGGAIATAWAAQLQPTYAPELRIAGAAAGGTPTDYALLYGSMNRGVGAGLFAAATIGQARE FPELVQIFGDFALYCAIRAKNMPQPPLAAAGLLRFDLDLLAAIAKPFESELGQHVIAANRPGALTPTMPV LLYHGSRSKAVGDLFIPEEGALALRDAWRANGADVDYWALPGEHVTADMFAIPWVVNWIRRKLLG*
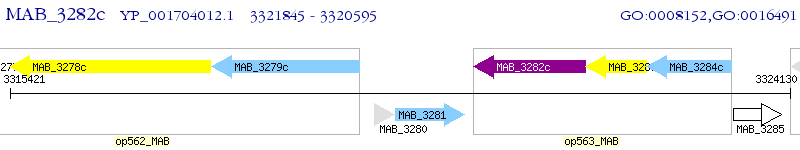
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3282c | - | - | 100% (416) | lipase |
| M. abscessus ATCC 19977 | MAB_4707c | - | e-150 | 63.95% (405) | putative lipase |
| M. abscessus ATCC 19977 | MAB_2676 | - | 3e-26 | 29.09% (385) | hypothetical protein MAB_2676 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1618c | - | 2e-28 | 28.87% (381) | hypothetical protein Mb1618c |
| M. gilvum PYR-GCK | Mflv_3621 | - | 1e-29 | 29.84% (382) | triacylglycerol lipase |
| M. tuberculosis H37Rv | Rv1592c | - | 2e-28 | 28.87% (381) | hypothetical protein Rv1592c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2389 | - | 8e-27 | 28.68% (380) | hypothetical protein MMAR_2389 |
| M. avium 104 | MAV_3194 | - | 1e-24 | 28.50% (407) | secretory lipase family protein |
| M. smegmatis MC2 155 | MSMEG_3197 | - | 8e-24 | 29.89% (368) | lipase |
| M. thermoresistible (build 8) | TH_2810 | - | 2e-28 | 29.76% (373) | PUTATIVE Triacylglycerol lipase |
| M. ulcerans Agy99 | MUL_1566 | - | 5e-27 | 28.68% (380) | hypothetical protein MUL_1566 |
| M. vanbaalenii PYR-1 | Mvan_2796 | - | 1e-30 | 29.14% (374) | triacylglycerol lipase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1618c|M.bovis_AF2122/97 ---------MVEPGNLAGATGA---------------EWIGR----PPHE
Rv1592c|M.tuberculosis_H37Rv ---------MVEPGNLAGATGA---------------EWIGR----PPHE
MMAR_2389|M.marinum_M MPKAMKGALMVKLGSLADARAA---------------EWIGR----PPHE
MUL_1566|M.ulcerans_Agy99 ---------MVKLGSLADARAA---------------GWIGR----PPHE
MAV_3194|M.avium_104 ---------MAELGNLAGTHGA---------------EWIAR----PPHE
MSMEG_3197|M.smegmatis_MC2_155 ----------MDLSSAAKAADA---------------EWIGR----APHE
Mflv_3621|M.gilvum_PYR-GCK ----------MDLGNVARSTGA---------------EWIGQ----PRHE
Mvan_2796|M.vanbaalenii_PYR-1 ----------MDLGNVARSTGA---------------EWIGQ----PFHE
TH_2810|M.thermoresistible__bu ----------MDLGSAAQSTGA---------------EWIGR----PTHE
MAB_3282c|M.abscessus_ATCC_199 --MRWSTGAFAALGGITAAVATGGTALRSPVVVDRLNDWAARRLTVQQRA
.. : : : * .: :
Mb1618c|M.bovis_AF2122/97 ELQRKVRPLLPSDDPFYFPPAGYQHAVPGTVLRSRDVELAFMGLIPQPVT
Rv1592c|M.tuberculosis_H37Rv ELQRKVRPLLPSDDPFYFPPAGYQHAVPGTVLRSRDVELAFMGLIPQPVT
MMAR_2389|M.marinum_M ELQRKVRPLLPSDDPFYDPPAGYHHAEPGTVLRSREVELAFLGLIPQSIT
MUL_1566|M.ulcerans_Agy99 ELQRKVRPLLPSDDPFYDPPAGYHHAEPGTVLRSREVELAFLGLIPQSIT
MAV_3194|M.avium_104 ELQRKVRPLLPSDDPFYQPPLGFQHAEPGTVLRSRDVELAFLGLIPQPIK
MSMEG_3197|M.smegmatis_MC2_155 ELDRDARPGLPGEDPFYLPPAGYHHAEPGTVLRSRDVELAFLGLVPQALH
Mflv_3621|M.gilvum_PYR-GCK ELRPRTRPVIPADDPFYEPPEGYEHARPGTVLRSRDVELAFLGLIPQKFS
Mvan_2796|M.vanbaalenii_PYR-1 PLRPRSRPMIPAKDDFYEPPAGFEHARPGTVLRSRDVELAFLGVIPQKFT
TH_2810|M.thermoresistible__bu ALR-RTRPILPERDPFYEPPPGFEHARPGTVLRSRDVELAFMGLIPQKFT
MAB_3282c|M.abscessus_ATCC_199 DPYAAILPTPISGDDFYGDPGDLGLLAPGEVVRADRVTPRLP---LRRAT
* * ** * . ** *:*: * : :
Mb1618c|M.bovis_AF2122/97 ATQLLYRTTNMYGNPEATVTTVIVPAE-LAPGQTCPLLSYQCAIDAMSSR
Rv1592c|M.tuberculosis_H37Rv ATQLLYRTTNMYGNPEATVTTVIVPAE-LAPGQTCPLLSYQCAIDAMSSR
MMAR_2389|M.marinum_M ATQLLYRTTDMYGNPEATVTTVILPAE-VAPGQTYPLLSYQCAIDAMSSR
MUL_1566|M.ulcerans_Agy99 ATQLLYRTTDMYGNPEATVTTVILPAE-VAPGQTYPLLSYQCAIDAMSSR
MAV_3194|M.avium_104 AVQLLYRTMDMNGEPEATVTTVIVPAD-AVPGRPCPLLSYQCAIDAVSSR
MSMEG_3197|M.smegmatis_MC2_155 ATQLLYRTTDRNGAPEAAVTTVIMPPD-ARAG--CPIVSYQCAIDAISAT
Mflv_3621|M.gilvum_PYR-GCK ATQLLYRTTDLYGQPQATVTTVLAPTE-RTDRRPLPIVSYQCAIDAVAGR
Mvan_2796|M.vanbaalenii_PYR-1 ATQLLYRTTDLNGEAQATVTTVVVPSE-RTSDGPLPIVSYQCAIDAVAGR
TH_2810|M.thermoresistible__bu ATQLLYRTTGFDGRPRATVTTVVVPAE-RDKNGRCPVVSYQCAIDAVTSR
MAB_3282c|M.abscessus_ATCC_199 MQRIMVRSTDTAGNPVPVTAALIEPERPWRGPGSRPVVVRNQAINSLGLK
::: *: . * . ...:::: * *:: : **:::
Mb1618c|M.bovis_AF2122/97 CFPSYALRRRAKALGSLTQMELLMISAALAEGWAVSVPDHEGPKGLWGSP
Rv1592c|M.tuberculosis_H37Rv CFPSYALRRRAKALGSLTQMELLMISAALAEGWAVSVPDHEGPKGLWGSP
MMAR_2389|M.marinum_M CFPSYALRRGAKAIGSLTQLELLLITAAVAEGWAVSVPDHEGVRGLWGTP
MUL_1566|M.ulcerans_Agy99 CFPSYALRRGAKAIGSLTQLELLLITAAVAEGWAVSVPDHEGVRGLWGTP
MAV_3194|M.avium_104 CFPSYALRRRAKALGSIGQFELFLITAAVAEGWAVSVPDHEGLRGLWGAP
MSMEG_3197|M.smegmatis_MC2_155 CFPSYALRRHAKVTGGLAQFELLLITAALAEGWAVSVPDHEGLDGMWGAP
Mflv_3621|M.gilvum_PYR-GCK CFPSYALRRGAKALGAVAQFEFLLIAAALAEGWAVSVPDHEGTTGMWGAP
Mvan_2796|M.vanbaalenii_PYR-1 CFPSYALRRGAKALGSFAQFEFLLVAAALAEGWAVSVPDHEGTSGMWGAP
TH_2810|M.thermoresistible__bu CFPSYALRRGARAVGALAQFEFLLIAAALAEGWAVSVPDHEGPDGSWGAP
MAB_3282c|M.abscessus_ATCC_199 FTPSYRLTH------LWYRDNPPMFPFLSAQNYAVLFPDHEGPRMSYAAG
*** * : : : :.. *:.:** .***** :.:
Mb1618c|M.bovis_AF2122/97 YEPGYRVLDGIRAALNSERVGLSPATPIGLWGYSGGGLASAWAAEACGEY
Rv1592c|M.tuberculosis_H37Rv YEPGYRVLDGIRAALNSERVGLSPATPIGLWGYSGGGLASAWAAEACGEY
MMAR_2389|M.marinum_M YEPGYRVLDGIRAALSAERLALPQSAPIGLWGYSGGGLASAWAAEMCGEY
MUL_1566|M.ulcerans_Agy99 YEPGYRVLDGIRAALSAERLALPQSAPIGLWGYSGGGLASAWAAEMCGEY
MAV_3194|M.avium_104 YEPGYRVLDGIRAALGSERLGLSPSAPIGLWGYSGGGLASAWAAETCAEY
MSMEG_3197|M.smegmatis_MC2_155 YEPGYHVLDGLRAALNSERLPLTDASPIGLWGYSGGGLASGWAAEMSGSY
Mflv_3621|M.gilvum_PYR-GCK FEPGYHVLDGIRAAVNHRPLGLAEDAPVGLWGYSGGGLASAWAAEVHDSY
Mvan_2796|M.vanbaalenii_PYR-1 HEPGYHVLDGVRAALNHGPLGLSPDAPVGLWGYSGGGLASAWAAEVQDAY
TH_2810|M.thermoresistible__bu LEPGYHILDGLRAALGYERLNLAVDAPVGLWGYSGGGLASAWAAEVCRDY
MAB_3282c|M.abscessus_ATCC_199 KMAGHAVLDSVRGMLSERPD--LAESPIVMHGYSGGAIATAWAAQLQPTY
.*: :**.:*. :. :*: : *****.:*:.***: *
Mb1618c|M.bovis_AF2122/97 APDLDIVGAVLGSPVGDLGHTFRRLNGTLLAGLPALVVAALQHSYPGLAR
Rv1592c|M.tuberculosis_H37Rv APDLDIVGAVLGSPVGDLGHTFRRLNGTLLAGLPALVVAALQHSYPGLAR
MMAR_2389|M.marinum_M APELNIVGAVLGSPVGDLGNTFRRLNGTLLAGLPAMVVAALGHAYPGLDR
MUL_1566|M.ulcerans_Agy99 APDLNIVGAVLGSPVGDLGNTFRRLNGTLLAGLPAMVVAALGHAYPGLDR
MAV_3194|M.avium_104 APELNVVGAVLGSPVGDLGNTFRRLNGSFLSGLPALVVAALAHIYPDLDR
MSMEG_3197|M.smegmatis_MC2_155 APELNIVGAVLGSPVGDLGHTFRRLNGTVFSGLPALVVAALADIYPGLNR
Mflv_3621|M.gilvum_PYR-GCK APELNVVGAVLGSPVGDLGHAFRRLNGSIYSGLPAMVVAALTHVYPDLNR
Mvan_2796|M.vanbaalenii_PYR-1 APELNVVGAVLGSPVGDLGHTFRRLNGSIYAGLPALVVAALTHIYPDLHR
TH_2810|M.thermoresistible__bu APDLNVVGAVLGSPVGDLGSAFRRLNGSLYAGLPAMVVAALAHIFPEVDQ
MAB_3282c|M.abscessus_ATCC_199 APELRIAGAAAGGTPTDYALLYGSMNRGVGAGLFAAATIGQAREFPELVQ
**:* :.**. *.. * . : :* . :** * .. . :* : :
Mb1618c|M.bovis_AF2122/97 VIKEHANDEGRQLLEQLTEMTTVDAVIRMAGRDMGDFLDEPLEDILSTPE
Rv1592c|M.tuberculosis_H37Rv VIKEHANDEGRQLLEQLTEMTTVDAVIRMAGRDMGDFLDEPLEDILSTPE
MMAR_2389|M.marinum_M VIKQHANEDGIELLERLEQMTTAEAVIRMANNDLGHYLDEPLDDVLSTPE
MUL_1566|M.ulcerans_Agy99 VIKQHANEDGIELLERLEQMTTAEAVIRMANNDLGHYLDEPLEDVLSTPE
MAV_3194|M.avium_104 VIKEHANEEGRALLDSLEKMTTVEAVIRMAGKNMGDYLDAPLEQILSTPE
MSMEG_3197|M.smegmatis_MC2_155 VIAEHATTEGRKLLQRLHKMSTIEAVLRLARKDMDDLVDLPLEQILDSPE
Mflv_3621|M.gilvum_PYR-GCK IIQQHATVEGKAMLARIEKMTTAHAMLSLVGKDMAHWVDRPLEEILLTPE
Mvan_2796|M.vanbaalenii_PYR-1 VIQEHATEDGKVMLARIEKMTTAHAMISLVGRDMAHMIDRPLEEILLSPE
TH_2810|M.thermoresistible__bu IIDEHATETGKAMLQRVEKMTTAHAVLRMIGMDMGKLVDQPLEEILQTPA
MAB_3282c|M.abscessus_ATCC_199 IFGDFALYCAIRAKNMPQPPLAAAGLLRFD-LDLLAAIAKPFESELG---
:: :.* . : .:: : :: : *::. *
Mb1618c|M.bovis_AF2122/97 VSHVFGDTKLGSAVPTPPVLIVQ-----AVHDYLIDVSDIDALADSYTAG
Rv1592c|M.tuberculosis_H37Rv ISHVFGDTKLGSAVPTPPVLIVQ-----AVHDYLIDVSDIDALADSYTAG
MMAR_2389|M.marinum_M VVHVFDDIKLGAAVPAPPVLIVQ-----AVHDYLISVDDIDTLAESYSTG
MUL_1566|M.ulcerans_Agy99 VVHVFDDIKLGAAVPAPPVLIVQ-----AVHDYLISVDDIDTLAESYSTG
MAV_3194|M.avium_104 VTYVFDHIRLGAAVPTPPVLIVQ-----AVHDYLIDVHDIDVLADAYSAG
MSMEG_3197|M.smegmatis_MC2_155 VTEVFDDIKLGVATPNPPVLLIQ-----AVHDELISVSSIDELAEKYLTG
Mflv_3621|M.gilvum_PYR-GCK VQHVFDSIKLGTSAPKVPVLIVQ-----AVHDRIISVDDIDELTETYTAG
Mvan_2796|M.vanbaalenii_PYR-1 VQHVFDSIKLGTAAPKVPLLIVQ-----AVHDRIISVDDIDALTETYTSG
TH_2810|M.thermoresistible__bu MQRVFDRIKLGSAIPAPPVLLVQ-----ACHDRIVSVDDIDTLADTYIEG
MAB_3282c|M.abscessus_ATCC_199 -QHVIAANRPGALTPTMPVLLYHGSRSKAVGDLFIPEEGALALRDAWRAN
*: : * * *:*: : * * :: . * : : .
Mb1618c|M.bovis_AF2122/97 GANVTYHRDLFSEHVSLHPLSAPMTLRWLTDRFAGKPLTDHRVRTTWPTI
Rv1592c|M.tuberculosis_H37Rv GANVTYHRDLFSEHVSLHPLSAPMTLRWLTDRFAGKPLTDHRVRTTWPTI
MMAR_2389|M.marinum_M GARVTYHRDAFNEHMMLHPLSAPMTLRWLTDRFAGLPLTEHVIRTTWPTM
MUL_1566|M.ulcerans_Agy99 GARVTYHRDAFNEHMMLHPLSAPMTLRWLTDRFAGLPLTEHVIRTTWPTM
MAV_3194|M.avium_104 GAQVTYHRDAFNEHMMLHPLSAPMTLRWLTDRFEGRPVTEHLIRTTWPTM
MSMEG_3197|M.smegmatis_MC2_155 GASVTYHRDLFSEHLLLHPMSAPMALRWLTDRFAARPLTANLVRSKWPTL
Mflv_3621|M.gilvum_PYR-GCK GASVTYHRDMFSEHLLLHPMSAPMTLRWLRDRFAGRPISAHIARTKWPTL
Mvan_2796|M.vanbaalenii_PYR-1 GASVTYHRDMLSEHLLLHPMSAPMTLRWLRDRFAGRPLRTHISRTKWPTL
TH_2810|M.thermoresistible__bu GVRVTYHRDMFSEHMLLHPMSAPMTLRWLRDRFAGQPLKHNLTRTKWPTM
MAB_3282c|M.abscessus_ATCC_199 GADVDYW-ALPGEHVTADMFAIPWVVNWIRRKLLG---------------
*. * * .**: . :: * .:.*: :: .
Mb1618c|M.bovis_AF2122/97 FNPMTYAGMARLAVIAAKVITGRKLSRRPL--------------------
Rv1592c|M.tuberculosis_H37Rv FNPMTYAGMARLAVIAAKVITGRKLSRRPL--------------------
MMAR_2389|M.marinum_M FNPMTYAGMARLAVIAAKMITGRKIHRRPL--------------------
MUL_1566|M.ulcerans_Agy99 FNPMTYAGMARLAVTAAKMITGRKIHRRPL--------------------
MAV_3194|M.avium_104 FNPMTYVGMARLLKIAAKVVTGRKIHRRPL--------------------
MSMEG_3197|M.smegmatis_MC2_155 LNPVTYVGMARLAGIAAKVVTGRALRRSPL--------------------
Mflv_3621|M.gilvum_PYR-GCK LNPSTYRGMLTLGKITAKVVLGRRVERQP---------------------
Mvan_2796|M.vanbaalenii_PYR-1 LNPSTYRGMLTLGKITAKVMLGRRVERQP---------------------
TH_2810|M.thermoresistible__bu FNPSTYRGMLRLGLITAKVLTGRRVDRRPXXXRLHAEMIAELRWPGDPDP
MAB_3282c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1618c|M.bovis_AF2122/97 --------------------------------------------------
Rv1592c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2389|M.marinum_M --------------------------------------------------
MUL_1566|M.ulcerans_Agy99 --------------------------------------------------
MAV_3194|M.avium_104 --------------------------------------------------
MSMEG_3197|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3621|M.gilvum_PYR-GCK ------------------------------LSQFDL--------------
Mvan_2796|M.vanbaalenii_PYR-1 ------------------------------LSRFDQ--------------
TH_2810|M.thermoresistible__bu DSGIDIWAMELDPGQLAVLDILRRPDVMTALTEWDAGDALGDYTRERIRS
MAB_3282c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1618c|M.bovis_AF2122/97 --------------------------------------------------
Rv1592c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2389|M.marinum_M --------------------------------------------------
MUL_1566|M.ulcerans_Agy99 --------------------------------------------------
MAV_3194|M.avium_104 --------------------------------------------------
MSMEG_3197|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3621|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2796|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2810|M.thermoresistible__bu SSALAVVTVEGREPADYLRGGSALEAVWIRAQQRGLAVHPVSPLFLYAHE
MAB_3282c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1618c|M.bovis_AF2122/97 --------------------------------------------------
Rv1592c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2389|M.marinum_M --------------------------------------------------
MUL_1566|M.ulcerans_Agy99 --------------------------------------------------
MAV_3194|M.avium_104 --------------------------------------------------
MSMEG_3197|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3621|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2796|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2810|M.thermoresistible__bu EDELRALSPRFASSLVNLRREFHDLAGTRRGESQIIVFRLFEAPPASVRS
MAB_3282c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1618c|M.bovis_AF2122/97 ----------
Rv1592c|M.tuberculosis_H37Rv ----------
MMAR_2389|M.marinum_M ----------
MUL_1566|M.ulcerans_Agy99 ----------
MAV_3194|M.avium_104 ----------
MSMEG_3197|M.smegmatis_MC2_155 ----------
Mflv_3621|M.gilvum_PYR-GCK ----------
Mvan_2796|M.vanbaalenii_PYR-1 ----------
TH_2810|M.thermoresistible__bu RRRSGGIGVD
MAB_3282c|M.abscessus_ATCC_199 ----------