For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRLNYLAAALAVAMTMLSGCAIDHSGLQNGKPTIRIGYQNFPSGDLVVKQNRWLETALPDYNIKWTRFDS GADINTAFIARELDFGALGSSPFARGLSAPLNIPYRVAFVLDVAGDNEALVASNRSGVSTIAGLKGKRVG TPFASTAHYSLLSALAQNGLSASDVRLVDLQPQAALAAFDRGDVDAVYTWLPTVDQVRKNGRDLITSRQL AADGRPTLDLAVVANAFADAHPEIVDAWRRQQARALRLIHDDPSAAAKAIAAENGLTPQEVADQLKQGIY LTPAEVGSAKWLGEQGKPGHIAVDLQSASQFLADQKQIPAAAPLATFENAVYTKGLPDAIGR
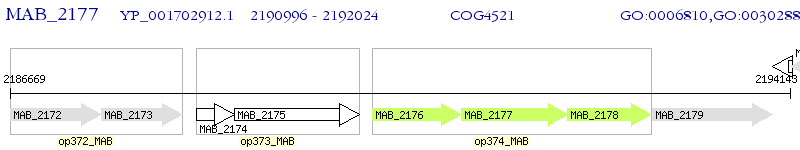
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2177 | - | - | 100% (342) | ABC transporter periplasmic protein |
| M. abscessus ATCC 19977 | MAB_2241 | - | 1e-24 | 28.18% (291) | putative nitrate ABC transporter, periplasmic protein |
| M. abscessus ATCC 19977 | MAB_2218 | - | 1e-13 | 27.11% (284) | sulfonate ABC transporter periplasmic sulfonate-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0723 | - | 1e-138 | 68.90% (344) | substrate-binding region of ABC-type glycine betaine transport |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3400 | - | 6e-22 | 26.13% (287) | ABC transporter substrate-binding protein |
| M. smegmatis MC2 155 | MSMEG_0114 | - | 1e-144 | 71.93% (342) | extracellular solute-binding protein, family protein 3 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0122 | - | 1e-143 | 71.43% (343) | substrate-binding region of ABC-type glycine betaine transport |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0723|M.gilvum_PYR-GCK -MRFKALLVALVAGMLAL-AGCSVGSGGQSGNDPEKPTIRIGYQTFPS--
Mvan_0122|M.vanbaalenii_PYR-1 -MKLKALLVVLVSAVLAL-AGCSVDNGGQHGDDSGKPTIRIGYQTFPS--
MSMEG_0114|M.smegmatis_MC2_155 -MKLKSLLVATAASALAL-AGCAVDNSEQ---DAEKPTIRVGYQTFPS--
MAB_2177|M.abscessus_ATCC_1997 -MRLNYLAAALAVAMTML-SGCAIDHSGLQ---NGKPTIRIGYQNFPS--
MAV_3400|M.avium_104 MRRHAALLASVLIAVAALGTGCSLESLSQS---AGVVNVVVGYQSKTINT
: * . . * :**:: .: :***. .
Mflv_0723|M.gilvum_PYR-GCK --GDLIVKNNKWLEEALPDHN--------IKWVKFDSGADVNTAFVADEL
Mvan_0122|M.vanbaalenii_PYR-1 --GDLIVKNNKWLEEALPDYN--------IKWTKFDSGADVNTAFVAGEL
MSMEG_0114|M.smegmatis_MC2_155 --GDLIVKNNGWLEEALPDYN--------IKWTKFDSGADVNTAFVAGEL
MAB_2177|M.abscessus_ATCC_1997 --GDLVVKQNRWLETALPDYN--------IKWTRFDSGADINTAFIAREL
MAV_3400|M.avium_104 VTAGTLLRAQGYLERRLADITTRTGTKYAVRWQDYDTGAPITAQMLAEKI
.. ::: : :** *.* . ::* :*:** :.: ::* ::
Mflv_0723|M.gilvum_PYR-GCK DFGALGSSPVARGLSEPLNIPYQVAFILDVAGDN-----EALVARNAAGI
Mvan_0122|M.vanbaalenii_PYR-1 DFGALGSSPVARGLSEPLNIPYKVAFVLDVAGDN-----EALVVRNGAGV
MSMEG_0114|M.smegmatis_MC2_155 DFGALGSSPVARGLSAPLNIPYKVAFVLDVAGDN-----EALVARDGSGV
MAB_2177|M.abscessus_ATCC_1997 DFGALGSSPFARGLSAPLNIPYRVAFVLDVAGDN-----EALVASNRSGV
MAV_3400|M.avium_104 DIGSMGDYPMLINGSKTQANPLARTEMVSITGYNPKGALNMVVVSPESRA
*:*::*. *. . * . * : ::.::* * : :*. :
Mflv_0723|M.gilvum_PYR-GCK TTIADLRDKRVATPFASTAHYSLLAALDQNGLSPNDVQLIDLQPQAILAA
Mvan_0122|M.vanbaalenii_PYR-1 DTIAQLKGRRIGTPFASTAHYSLLAALDQNGLSANDVQLIDLQPQAILAA
MSMEG_0114|M.smegmatis_MC2_155 NSIADLRGKRVATPFASTAHYSLLAALAQNGLSPNDVQLIDLQPQAILAA
MAB_2177|M.abscessus_ATCC_1997 STIAGLKGKRVGTPFASTAHYSLLSALAQNGLSASDVRLVDLQPQAALAA
MAV_3400|M.avium_104 KDLADLAGAKVSASVGSAGHGTLVRALSRAGVN--GVEVLNQQPQVGASA
:* * . ::.:...*:.* :*: ** : *:. .*.::: ***. :*
Mflv_0723|M.gilvum_PYR-GCK WERGDIDAAYTWLPTLNELRKTGKDLITSRQLAAEGKPTLDLAVVSSEFA
Mvan_0122|M.vanbaalenii_PYR-1 WERGDIDAAYTWLPTLDELRKTGRDLITSRQLADAGKPTLDLATVSDEFA
MSMEG_0114|M.smegmatis_MC2_155 WERGDIAAAYSWLPTLDDLRKTGKDLITSRELAADGKPTLDLGVVSDKFA
MAB_2177|M.abscessus_ATCC_1997 FDRGDVDAVYTWLPTVDQVRKNGRDLITSRQLAADGRPTLDLAVVANAFA
MAV_3400|M.avium_104 LESGQVQALSQFVAWPGLLVFQGKAKLLYDGAELN-LPTLHGVVVRRSYA
: *:: * ::. . : *: : ***. .* :*
Mflv_0723|M.gilvum_PYR-GCK KSHPEVVDTWRKQQARALDVIADDPAAAAKAIAAEVGLTPE---------
Mvan_0122|M.vanbaalenii_PYR-1 SAHPEAVDVWRQQQGRALDLIREDPQAAAEAIAAEIGLTPQ---------
MSMEG_0114|M.smegmatis_MC2_155 TEHPDVVDTWREQEARALDVIKDDPDAAAKAIAAEIGLSPE---------
MAB_2177|M.abscessus_ATCC_1997 DAHPEIVDAWRRQQARALRLIHDDPSAAAKAIAAENGLTPQ---------
MAV_3400|M.avium_104 AAHPEVLAAFLQAQLDATDFLNTHPLQAARIVADASGLPPEVVYLYNGPG
**: : .: . : * .: .* **. :* **.*:
Mflv_0723|M.gilvum_PYR-GCK ----------DIEGQLTQTVFLTP-----EQVASAEWLGDD---------
Mvan_0122|M.vanbaalenii_PYR-1 ----------DVAGQLKQMVFLTP-----QDISSTEWLGTE---------
MSMEG_0114|M.smegmatis_MC2_155 ----------DVAGQLKQGVYLTP-----AEVASEQWLGSD---------
MAB_2177|M.abscessus_ATCC_1997 ----------EVADQLKQGIYLTP-----AEVGSAKWLGEQ---------
MAV_3400|M.avium_104 GTSFDTTLKPSLTEALKSDVPYLKSIGDFADLDVDKFVVDEPLRAVFTAR
.: *.. : :: ::: :
Mflv_0723|M.gilvum_PYR-GCK -----------GKPGNLAANLQSASEFLAE-----QKQIPAAAPLNTFQD
Mvan_0122|M.vanbaalenii_PYR-1 -----------GNPGNLAVNLESASQFLAD-----QSQIPAAAPLKTFQD
MSMEG_0114|M.smegmatis_MC2_155 -----------GKPGNIAANLQSASEFLAE-----QKQIPEAAPLGTFED
MAB_2177|M.abscessus_ATCC_1997 -----------GKPGHIAVDLQSASQFLAD-----QKQIPAAAPLATFEN
MAV_3400|M.avium_104 GLDYQAARARTTNPSTLRGDPALAGELWLDGADTTQTTADPASLLRAVRD
:*. : : *.:: : *. *: * :..:
Mflv_0723|M.gilvum_PYR-GCK GL---------------------------------------------YTR
Mvan_0122|M.vanbaalenii_PYR-1 AV---------------------------------------------YTK
MSMEG_0114|M.smegmatis_MC2_155 AI---------------------------------------------YTK
MAB_2177|M.abscessus_ATCC_1997 AV---------------------------------------------YTK
MAV_3400|M.avium_104 ALGRGARVRAAYVPDTEFGTRWFADKAFWVKDGQNYLPFGTAAGAGRYLA
.: *
Mflv_0723|M.gilvum_PYR-GCK GLPGVLTE----------
Mvan_0122|M.vanbaalenii_PYR-1 GLPGALNE----------
MSMEG_0114|M.smegmatis_MC2_155 GLPGAISK----------
MAB_2177|M.abscessus_ATCC_1997 GLPDAIGR----------
MAV_3400|M.avium_104 AHPGSIAVNYQQALGGSV
. *. :