For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RRHAALLASVLIAVAALGTGCSLESLSQSAGVVNVVVGYQSKTINTVTAGTLLRAQGYLERRLADITTRT GTKYAVRWQDYDTGAPITAQMLAEKIDIGSMGDYPMLINGSKTQANPLARTEMVSITGYNPKGALNMVVV SPESRAKDLADLAGAKVSASVGSAGHGTLVRALSRAGVNGVEVLNQQPQVGASALESGQVQALSQFVAWP GLLVFQGKAKLLYDGAELNLPTLHGVVVRRSYAAAHPEVLAAFLQAQLDATDFLNTHPLQAARIVADASG LPPEVVYLYNGPGGTSFDTTLKPSLTEALKSDVPYLKSIGDFADLDVDKFVVDEPLRAVFTARGLDYQAA RARTTNPSTLRGDPALAGELWLDGADTTQTTADPASLLRAVRDALGRGARVRAAYVPDTEFGTRWFADKA FWVKDGQNYLPFGTAAGAGRYLAAHPGSIAVNYQQALGGSV*
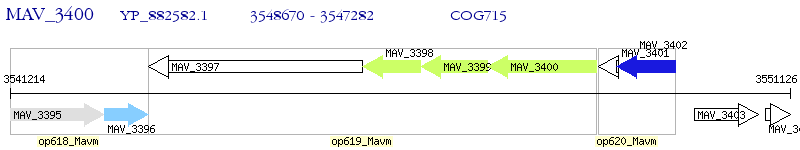
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3400 | - | - | 100% (462) | ABC transporter substrate-binding protein |
| M. avium 104 | MAV_0141 | - | 3e-13 | 22.96% (331) | sulfonate binding protein |
| M. avium 104 | MAV_2435 | - | 3e-09 | 24.89% (237) | sulfonate binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0723 | - | 5e-20 | 27.74% (292) | substrate-binding region of ABC-type glycine betaine transport |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2241 | - | 1e-179 | 70.17% (466) | putative nitrate ABC transporter, periplasmic protein |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1232 | - | 1e-115 | 50.21% (478) | ABC transporter substrate-binding protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0122 | - | 6e-20 | 27.68% (289) | substrate-binding region of ABC-type glycine betaine transport |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3400|M.avium_104 ------------MRRHAALLASVLIAVAALGTGCSLESLSQSAGVVNVVV
MAB_2241|M.abscessus_ATCC_1997 ------------MKR-TALLASMLIALA---TGCSLDSGSGSSQEVTVVI
MSMEG_1232|M.smegmatis_MC2_155 MDEAQQDSGGRWRARSAALLAAVTVGGAQLLTGCGLLTEP----TVVVNV
Mflv_0723|M.gilvum_PYR-GCK --------------MRFKALLVALVAGMLALAGCSVGSGG--------QS
Mvan_0122|M.vanbaalenii_PYR-1 --------------MKLKALLVVLVSAVLALAGCSVDNGG--------QH
* :. :**.: .
MAV_3400|M.avium_104 GYQSKTINTVTAGTLLRAQGYLERRLADITTRTGTKYAVRWQDYDTGAPI
MAB_2241|M.abscessus_ATCC_1997 GYQSKTINTVTAGTLLRAQGYLEKRLGDLTSRTGTKYTVQWQDYDTGAPI
MSMEG_1232|M.smegmatis_MC2_155 GYQSKTINTVNAGTLLRDRGDFEAELKQIGAETGTKYRVVWQDFASGAPL
Mflv_0723|M.gilvum_PYR-GCK GNDPEKP-TIRIGYQTFPSGDLIVKNNKWLEEALPDHNIKWVKFDSGADV
Mvan_0122|M.vanbaalenii_PYR-1 GDDSGKP-TIRIGYQTFPSGDLIVKNNKWLEEALPDYNIKWTKFDSGADV
* :. . *: * * : . . .: ..: : * .: :** :
MAV_3400|M.avium_104 TAQMLAEKIDIGSMGDYPMLINGSKTQANPLARTEMVSITGYNPKGALNM
MAB_2241|M.abscessus_ATCC_1997 TAQMLAEKIDIGSMGDYPLLINGSKTQANEKARTELVSITGYNAKGSLNM
MSMEG_1232|M.smegmatis_MC2_155 TAQMIATHVDIGSMGDYPLLTNGSKTRKYDDAATEMVAITGYNLRGSLNQ
Mflv_0723|M.gilvum_PYR-GCK NTAFVADELDFGALGSSPVARGLSEPLNIPYQVAFILDVAGDN-----EA
Mvan_0122|M.vanbaalenii_PYR-1 NTAFVAGELDFGALGSSPVARGLSEPLNIPYKVAFVLDVAGDN-----EA
.: ::* .:*:*::*. *: . *:. : :: ::* * :
MAV_3400|M.avium_104 VVVSPESRAKDLADLAGAKVSASVGSAGHGTLVRALSRAGVN---GVEVL
MAB_2241|M.abscessus_ATCC_1997 VVVPPNSAITELPELKGKKVSASVGSAGHGTLVRALRNDGLDPTRDVEVL
MSMEG_1232|M.smegmatis_MC2_155 VVVPSDSSAQKLEDLTGKKVSTSLGSAGDGMFSSALKGNGIDKN-AVQIV
Mflv_0723|M.gilvum_PYR-GCK LVARNAAGITTIADLRDKRVATPFASTAHYSLLAALDQNGLSPN-DVQLI
Mvan_0122|M.vanbaalenii_PYR-1 LVVRNGAGVDTIAQLKGRRIGTPFASTAHYSLLAALDQNGLSAN-DVQLI
:*. : : :* . ::.:...*:.. : ** *:. *:::
MAV_3400|M.avium_104 NQQPQVGASALESGQVQALSQFVAWPGLLVFQGKAKLLY-DGAELNLPTL
MAB_2241|M.abscessus_ATCC_1997 NQQPQVGASALESGQVQALSQFVAWPGLLVFQNKAKLLY-DGAELNVPTF
MSMEG_1232|M.smegmatis_MC2_155 NQDPSVGASAIDGRQVDALAQFVPWPQLVIYRNQGRLLY-DGGDNNVPTF
Mflv_0723|M.gilvum_PYR-GCK DLQPQAILAAWERGDIDAAYTWLPTLNELRKTGKDLITSRQLAAEGKPTL
Mvan_0122|M.vanbaalenii_PYR-1 DLQPQAILAAWERGDIDAAYTWLPTLDELRKTGRDLITSRQLADAGKPTL
: :*.. :* : :::* ::. : .: : : . . **:
MAV_3400|M.avium_104 HGVVVRRSYAAAHPEVLAAFLQAQLDATDFLNTHPLQAARIVADASGLPP
MAB_2241|M.abscessus_ATCC_1997 HGVVVRRDYATQHPEVLDAFLQAQLDATDFIHEKSLEAARIVAEGSGLPQ
MSMEG_1232|M.smegmatis_MC2_155 HGVVVRRQFADQRPEVMAAFLRAMRSTTDAIVADPLGAAQRIGELTGIEP
Mflv_0723|M.gilvum_PYR-GCK DLAVVSSEFAKSHPEVVDTWRKQQARALDVIADDPAAAAKAIAAEVGLTP
Mvan_0122|M.vanbaalenii_PYR-1 DLATVSDEFASAHPEAVDVWRQQQGRALDLIREDPQAAAEAIAAEIGLTP
. ..* .:* :**.: .: : : * : .. **. :. *:
MAV_3400|M.avium_104 EVVYLYNGP-GGTSFDTTLKPSLTEALKSDVPYLKSIGDFAD-LDVDKFV
MAB_2241|M.abscessus_ATCC_1997 EVVYLYNGP-GGTSFDATLKPSLIEAFTSDVPYLKSIGDFAD-LNIDDFV
MSMEG_1232|M.smegmatis_MC2_155 EVIYLYNGPNGLVSFDMTLKQQFLGAFAAVKDYLVGRGSVTENFDIPGFV
Mflv_0723|M.gilvum_PYR-GCK EDIEG------QLTQTVFLTPEQVASAEWLGDDGKPGN------------
Mvan_0122|M.vanbaalenii_PYR-1 QDVAG------QLKQMVFLTPQDISSTEWLGTEGNPGN------------
: : . *. . : .
MAV_3400|M.avium_104 VDEPLRAVFTARGLDYQAARARTTNPSTLR----------GDPALAGELW
MAB_2241|M.abscessus_ATCC_1997 HDGPLRQAYTTRAKNYETELAAMVNPLVLHPA------DGADPGQAAEIW
MSMEG_1232|M.smegmatis_MC2_155 NDSYLREMFGD---EYPQRSASMANPMTLSGFDEICRLPVDDPAQGSELW
Mflv_0723|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0122|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3400|M.avium_104 LDGADTTQTTADPASLLRAVRDALGRGARVRAAYVPDTEFGTRWFADKAF
MAB_2241|M.abscessus_ATCC_1997 FDGKDSTQVYPTAQELLKAVNAANAAGRKVRAAYVADTELGTRWFADHAR
MSMEG_1232|M.smegmatis_MC2_155 AAGSDTTDVAATPSCLLRRVAATS----EVRAAYVPDTLTGLRIFADHAV
Mflv_0723|M.gilvum_PYR-GCK ---------------------------------LAANLQSASEFLAEQKQ
Mvan_0122|M.vanbaalenii_PYR-1 ---------------------------------LAVNLESASQFLADQSQ
. : . . :*::
MAV_3400|M.avium_104 WVKDGQN-----YLPFGTAAGAGRYLAAHPGSIAVNYQQALGGSV---
MAB_2241|M.abscessus_ATCC_1997 WVQDPAG-----LHPFTTGAGAARYVAAHPGARPLDYAQALAAAS---
MSMEG_1232|M.smegmatis_MC2_155 WLQDPSAPPTHRYKPFATSAGAESYRAEHPDADAVSYTAAVGHSRAAR
Mflv_0723|M.gilvum_PYR-GCK IPAAAPLN----------TFQDGLYTRGLPGVLTE-------------
Mvan_0122|M.vanbaalenii_PYR-1 IPAAAPLK----------TFQDAVYTKGLPGALNE-------------
* *.