For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLNRRDSHHDLDGAVFSRRQLLVRGGAALALVAVLTAGLLIKSTGVLDRYVNVIADLHNVGDGLPPRSDV KYRGVLVGAVDGVTPSAGTQPNRVRVRLKPMYARSIPATVTARVVPSNVFAVSALQLVDHGPGAPIREGG HIAEDTRLPTVLFQTTVNKLRQILTAAGRGREDNTVGILAAVGAATNHRRAELLTSAAQLDRMLDQLNAI VATDQGPSTVSALVQAAHGLSQTAPDLVDALHQAVRPMQTLAEKREQLRTFIGAGVQTTGTTVRSLHNHT DQLIQITTDLTPVLGVLADNAQHFVPITRRITRFSDTFFREVWDPELATAKGRVNLSFTPSYTYTRADCP RYGELKGPSCFTAPQIAVRPDLPEVLLPQNYQPPANLAPPPGTVLGDNGNLIAVGPPLVNPNPDLRDPNP PVPPWLTPAPPVPGSADPGQILTAPASFGGNVGPVGSTQERGQLGQILGQPATSADQLVLGPLARGTVVT RAADPHEEGGAR
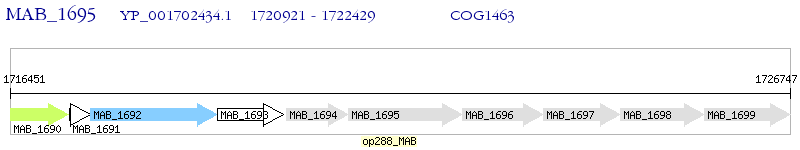
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1695 | - | - | 100% (502) | putative Mce family protein |
| M. abscessus ATCC 19977 | MAB_1010c | - | e-171 | 61.54% (494) | putative MCE family protein |
| M. abscessus ATCC 19977 | MAB_4568c | - | e-171 | 62.12% (491) | putative Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3526c | mce4D | 2e-09 | 21.58% (468) | MCE-family protein MCE4D |
| M. gilvum PYR-GCK | Mflv_0010 | - | 1e-165 | 57.30% (541) | hypothetical protein Mflv_0010 |
| M. tuberculosis H37Rv | Rv3496c | mce4D | 2e-09 | 21.58% (468) | MCE-family protein MCE4D |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0177 | mce6A | 0.0 | 68.87% (485) | MCE-family protein Mce6A |
| M. avium 104 | MAV_0102 | - | 1e-175 | 61.45% (511) | mce related protein |
| M. smegmatis MC2 155 | MSMEG_1143 | - | 1e-173 | 59.81% (520) | mce related protein |
| M. thermoresistible (build 8) | TH_2386 | - | 1e-162 | 63.24% (438) | mce related protein |
| M. ulcerans Agy99 | MUL_4919 | mce6A | 0.0 | 68.45% (485) | MCE-family protein Mce6A |
| M. vanbaalenii PYR-1 | Mvan_0959 | - | 1e-176 | 60.58% (520) | hypothetical protein Mvan_0959 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0177|M.marinum_M MTN----HIDLDGIYFSKRQLLARGGIALVVVLLVTSWLLAKSMGYLDRG
MUL_4919|M.ulcerans_Agy99 MTN----HIDLDGIYFSKRQLLARGGIALVVVLLVTSWLLAKSMGYLDRG
MAB_1695|M.abscessus_ATCC_1997 MLNRRDSHHDLDGAVFSRRQLLVRGGAALALVAVLTAGLLIKSTGVLDRY
MSMEG_1143|M.smegmatis_MC2_155 MGN----SIELDGRGPSERQLLACGLAVLVVAGLISTVLLVKATGRLDPY
TH_2386|M.thermoresistible__bu MAN----SFDLDGRGPTDRQLLAGGVVVIVVLSVISTYLLVKATGRLDPF
Mflv_0010|M.gilvum_PYR-GCK MAN----SFEFDGRGPSDRQLLVCGVAVLAIVSIVTAVLLVKSTGRLDPR
Mvan_0959|M.vanbaalenii_PYR-1 MAN----SLEMDGRGPTDRQLLACGVALLVIALVVSGALLVKSTGRMDAH
MAV_0102|M.avium_104 MAN----SLDFDGRGPTDHQLLALGATVLLIAGLITGGLLLKSTGRLNDY
Mb3526c|M.bovis_AF2122/97 MMG--------RVAMLTGSRGLRYATVIALVAALVGGVYVLSSTG----N
Rv3496c|M.tuberculosis_H37Rv MMG--------RVAMLTGSRGLRYATVIALVAALVGGVYVLSSTG----N
* . : : * . : :: : .: *
MMAR_0177|M.marinum_M VQVVADLRNIGDGLPAGSDVKYRGIQVGTVESLDPSIGNQPNQVRIRLKP
MUL_4919|M.ulcerans_Agy99 VQVVADLRNIGDGLPAGSDVKYRGIQVGTVESLDPSIGNQPNQVRIRLKP
MAB_1695|M.abscessus_ATCC_1997 VNVIADLHNVGDGLPPRSDVKYRGVLVGAVDGVTPSAGTQPNRVRVRLKP
MSMEG_1143|M.smegmatis_MC2_155 VRVVAALINVGDGLPQRSDVKYHGVLVGMVEDVTPAAHGEPNYVHINLNP
TH_2386|M.thermoresistible__bu VRVVADLVNVGDGLPQRSDVKYHGVLVGHVDSVTPAAHGHPNYVHINLDR
Mflv_0010|M.gilvum_PYR-GCK VRVVAAMVNVGDGLPQRSDVKYHGVLVGAVDDVTPAENGEPNFVHIDLKP
Mvan_0959|M.vanbaalenii_PYR-1 VRVVAALVNVGDGLPQRSDVKYHGVLVGAVDDVTPAQYGDPNFVHIDLKP
MAV_0102|M.avium_104 IRVVAELTNVGDGLPARSDVKYHGLLVGAVDNVIPAAYGKPNYVHINLKP
Mb3526c|M.bovis_AF2122/97 KRTIVGYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSD----VKITMSV
Rv3496c|M.tuberculosis_H37Rv KRTIVGYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSD----VKITMSV
..:. . . ** .:*: *: ** :: : * *:: :.
MMAR_0177|M.marinum_M EYAASIPVTVTARVVPSNVFAVSSIQLVEHGPGA--PISDGERISEDTDL
MUL_4919|M.ulcerans_Agy99 EYAASIPVTVTARVVPSNVFAVSSIQLVEHGPGT--PISDGERISEDTDL
MAB_1695|M.abscessus_ATCC_1997 MYARSIPATVTARVVPSNVFAVSALQLVDHGPGA--PIREGGHIAEDTRL
MSMEG_1143|M.smegmatis_MC2_155 EYASSIPSTVTARVVPSNVFAVSSVQLVDRDGGAGPAISAGAQIPEDTEL
TH_2386|M.thermoresistible__bu HHARSIPATVTARVVPSNVFAVSSVQLVDRDGGPGTPIQAGAHIPEDTEL
Mflv_0010|M.gilvum_PYR-GCK EYAKSIPANVTARVVPSNVFAVSSVQLVG-SGGP--PIQPGAEIPEDTEL
Mvan_0959|M.vanbaalenii_PYR-1 EYAASIPAGVTARVVPSNVFAVSSVQLVDRSGGP--PIQSGARIPEDTEL
MAV_0102|M.avium_104 EYAQDIPSAVTARVVPSNVFAVSSVQLVDGAPGP--SIRNGARIPEDLQL
Mb3526c|M.bovis_AF2122/97 SKDVKVPVDVQAVIMSPNLVAARFIQLTPVYTGG-AVLPDNGRIDLDRTA
Rv3496c|M.tuberculosis_H37Rv SKDVKVPVDVQAVIMSPNLVAARFIQLTPVYTGG-AVLPDNGRIDLDRTA
.:* * * ::..*:.*. :**. * : . .* *
MMAR_0177|M.marinum_M PTVLFQTALTKLRDILAAAGRNRDDHTLGVLAAVGAATNNRNGALQLSTH
MUL_4919|M.ulcerans_Agy99 PTVLFQTALTKLRDILAAAGRNRDDHTLGVLAAVGAATNNRNGALQLSTH
MAB_1695|M.abscessus_ATCC_1997 PTVLFQTTVNKLRQILTAAGRGREDNTVGILAAVGAATNHRRAELLTSAA
MSMEG_1143|M.smegmatis_MC2_155 PTVLFQTTISKLRDILAATGRGREDRTVGVLAAVNAATEGRRTELLTSGA
TH_2386|M.thermoresistible__bu PTVLFQTTISKLRDILAATGRGREDRTVGILAAVNAATENRRTELLSAGA
Mflv_0010|M.gilvum_PYR-GCK PTVLFQTTISKLRDILAATGRGREDDTVGILAAVHAATEDRRTELLTSGA
Mvan_0959|M.vanbaalenii_PYR-1 PTVLFQTTISKLRDILAATGRGREDDTVGILAAVNAATEDRRTELLTSGA
MAV_0102|M.avium_104 STVIFQTTISKLRDILAATGRGREDHSVGILAAVAAATNNRRGPLLTAGA
Mb3526c|M.bovis_AF2122/97 VPVEWDEVKEGLTRLAADLSPAAGELQGPLGAAINQAADTLDGNGDSLHN
Rv3496c|M.tuberculosis_H37Rv VPVEWDEVKEGLTRLAADLSPAAGELQGPLGAAINQAADTLDGNGDSLHN
.* :: . * : : . : : **: *::
MMAR_0177|M.marinum_M QLIRIIDELNGVVATDQ-GPSTVAALVDAARGLSQSAPELLDAFHDAVAP
MUL_4919|M.ulcerans_Agy99 QLIRIIDELNGVVATDE-GPSTVAALVDAARGLSQSAPELLDALHDAVAP
MAB_1695|M.abscessus_ATCC_1997 QLDRMLDQLNAIVATDQ-GPSTVSALVQAAHGLSQTAPDLVDALHQAVRP
MSMEG_1143|M.smegmatis_MC2_155 QLNRLIDEVDSIVATDPSAPTTVSTLIEATRGLQQTAPELVDALHTAVRP
TH_2386|M.thermoresistible__bu QLDLLIDEIDSIVATDP-GPTTVSALIGATRGLQQTAPELLDALHQAVQP
Mflv_0010|M.gilvum_PYR-GCK QLNRMVDELDRIVATEP-GPTTVSALVDATRGLQQTAPELVDALHQAVQP
Mvan_0959|M.vanbaalenii_PYR-1 QLNSLVDELDRIVATDP-GPTTVSALVDATRGLQQTAPELVDALHQAVEP
MAV_0102|M.avium_104 QLTRVLDELNAIVATDP-GPSTVSALLDATRGLQSTAPDLVDVLHDAVRP
Mb3526c|M.bovis_AF2122/97 ALRELAQVAGRLGDSRGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASV
Rv3496c|M.tuberculosis_H37Rv ALRELAQVAGRLGDSRGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASV
* : : . : : ** * . .*..: :::: ..
MMAR_0177|M.marinum_M MRTLVDKQQALHDFLGAGQYTTGLAVNSLQNHSDQLVQITTDLTPVVGVF
MUL_4919|M.ulcerans_Agy99 MRTLVDKQQALHDFLGAGQYTTGLAANSLQNHSDQLVQITTDLTPVVGVF
MAB_1695|M.abscessus_ATCC_1997 MQTLAEKREQLRTFIGAGVQTTGTTVRSLHNHTDQLIQITTDLTPVLGVL
MSMEG_1143|M.smegmatis_MC2_155 MQTLVEQRSQLDSLISAGMNTLDTTRTALNNHTDRLVKITSELTPVVGNL
TH_2386|M.thermoresistible__bu MQTLVQQRAQLDALIGGALHTSATTHTALTNHTDRMVTITTEMTPVIGNL
Mflv_0010|M.gilvum_PYR-GCK MQTLVEQSAQLTSLVNGGIRTSETTRTAFDNHADRLVRITTELTPVVGSL
Mvan_0959|M.vanbaalenii_PYR-1 MQTLVEQSAQLTTFINGGINTVGTTHTALNNHTDRMVRITTELTPVIGNL
MAV_0102|M.avium_104 MQTLVEKREQFRSLVTGSYHTFSVNRQAFDNHTDQLIEMTQNLTPVLGVF
Mb3526c|M.bovis_AF2122/97 SQVLADSSANLDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQTL
Rv3496c|M.tuberculosis_H37Rv SQVLADSSANLDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQTL
:.*.:. : : : : :: . :: . ::. . :
MMAR_0177|M.marinum_M ADNAQHFVPITERITRFSDKFFEEVWDPASTVAHGRVNVS-FTPSYTYTR
MUL_4919|M.ulcerans_Agy99 ADNAQHFVPITERITRFSDKFFEEVWDPASTVAHGRVNVS-FTPSYTYTR
MAB_1695|M.abscessus_ATCC_1997 ADNAQHFVPITRRITRFSDTFFREVWDPELATAKGRVNLS-FTPSYTYTR
MSMEG_1143|M.smegmatis_MC2_155 ADTSHHWLPAFVKLNNLSDKFFDEVWRPERNVGNMRMNLS-FTPSYTYTR
TH_2386|M.thermoresistible__bu ADTSHHWLPAFIKMNGLSDKFFEEVWRPERDIGNMRMNLS-LTPSYTYTR
Mflv_0010|M.gilvum_PYR-GCK ADTSRHWVPAFVKLNELSRKFFDEVWIHEHDIGNMRINLS-FTPTYSYTR
Mvan_0959|M.vanbaalenii_PYR-1 AQTSHHWVPAFVKLNGLSRKFFDEVWMHDRDVGNMRVNLS-FTPTYSYTR
MAV_0102|M.avium_104 AMNSDKFVPIFTRLNRLSDKFFQEVWDPELDTGNMRVNLA-LTPTYTYTR
Mb3526c|M.bovis_AF2122/97 SDQSE-NIEQVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFIC
Rv3496c|M.tuberculosis_H37Rv SDQSE-NIEQVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFIC
: : : :: . . * ::: : :.:. :: . :
MMAR_0177|M.marinum_M ADCPRYGELRAPSCFTAPLVQARPDLPEVLLPQNYQPPPNLAPPPGTVVG
MUL_4919|M.ulcerans_Agy99 ADCPRYGELRAPSCFTAPLVQARPDLPEVLLPQNYQPPPNLAPPPGTVVG
MAB_1695|M.abscessus_ATCC_1997 ADCPRYGELKGPSCFTAPQIAVRPDLPEVLLPQNYQPPANLAPPPGTVLG
MSMEG_1143|M.smegmatis_MC2_155 ADCPQYGELKGPSCFTAPLVVTRPALPDQMLPRNYQPPKDLAPPPGTVVG
TH_2386|M.thermoresistible__bu ADCPRYGELKGPSCYTAPLVVTRPSLPDVLLPQNYQPPKDLAPPPGTIVG
Mflv_0010|M.gilvum_PYR-GCK ADCPQYAGLKGPSCYTAPLVPTRAALPDVLLPQNYQPPKDLQPPPGTEIG
Mvan_0959|M.vanbaalenii_PYR-1 ADCPQYGELKGPSCFTAPLVPTRPELPDVLLPQNYQPPADLKPPPGTEVG
MAV_0102|M.avium_104 ADCPRYGQLQGPSCFTAPTIAVRPDLPEVLLPQNYHPPTDLAPPPGTQIG
Mb3526c|M.bovis_AF2122/97 GGSFDTAAGPSAPDYYRRAEICRERLGPVLRRLTVNYPPIMFHPLNTITA
Rv3496c|M.tuberculosis_H37Rv GGSFDTAAGPSAPDYYRRAEICRERLGPVLRRLTVNYPPIMFHPLNTITA
... . ... : * * : . : * : * .* .
MMAR_0177|M.marinum_M PDGNLVAVGPPLINPTPNLEDPNPPLPAG---VTPAPPVPGTANPDLLP-
MUL_4919|M.ulcerans_Agy99 PDGNLVAVGPPLINPTPNLEDPNPPLPAG---VTPAPPVPGTANPDLLP-
MAB_1695|M.abscessus_ATCC_1997 DNGNLIAVGPPLVNPNPDLRDPNPPVPPW---LTPAPPVPGSADPGQIL-
MSMEG_1143|M.smegmatis_MC2_155 ENGNLVAVGPPLINPNPSLADPNPPLPPW---MAPAPPVPGTANSALPVT
TH_2386|M.thermoresistible__bu ENGNLVAVGPPLVNPNPNLADPNPPLPPG---MFPAPPVPGTSNPDLVPI
Mflv_0010|M.gilvum_PYR-GCK PSGNLIAVGPPLVNPNPSLADPNPPLPPW---MPPSPPVPLTANPALVPP
Mvan_0959|M.vanbaalenii_PYR-1 PNGNLVAVGPPLVNPNPDLRDPNPPLPPW---LSPSPPVPGTANPALVPV
MAV_0102|M.avium_104 PDGNLVATGPPLYNPDPSLADPNPPLPWWPWQIGPAPRVPGTADPDDAPP
Mb3526c|M.bovis_AF2122/97 YKGQIIYDTPATEAKSETPVPELTWVPAG-----GGAPVGNPADLQSLLV
Rv3496c|M.tuberculosis_H37Rv YKGQIIYDTPATEAKSETPVPELTWVPAG-----GGAPVGNPADLQSLLV
.*::: *. . :* .. * .::
MMAR_0177|M.marinum_M ------------AAPSS---------------------------------
MUL_4919|M.ulcerans_Agy99 ------------AAPSS---------------------------------
MAB_1695|M.abscessus_ATCC_1997 ------------TAPAS---------------------------------
MSMEG_1143|M.smegmatis_MC2_155 PPPGVNQSPLAPVAPKPGSPPPPPPAPG-----PLPAEAAPA--------
TH_2386|M.thermoresistible__bu PPPPVNLSPLAPVAPNPNKPRPPVPAPP-----PAP--------------
Mflv_0010|M.gilvum_PYR-GCK PQLPAPLSPPAPVAPKPGDPPPRPAAPAPAAPAPAPAAPAPAPAAPGPGS
Mvan_0959|M.vanbaalenii_PYR-1 PQPPVPLSPPAPVAPKPGDPPPPPAAPPVSGPAPLPAEAAPA--------
MAV_0102|M.avium_104 PPPPS-----PPPSPAPPGPVAPA--------------------------
Mb3526c|M.bovis_AF2122/97 PPAPG-------PAPAPPAP------------------------------
Rv3496c|M.tuberculosis_H37Rv PPAPG-------PAPAPPAP------------------------------
:* .
MMAR_0177|M.marinum_M ---------FGGNVGPVGSVQERAALSLITGE--QATVATQLLLGPVARG
MUL_4919|M.ulcerans_Agy99 ---------FGGNVGPVGSVQERAALSLITGE--QATVATQLLLGPVARG
MAB_1695|M.abscessus_ATCC_1997 ---------FGGNVGPVGSTQERGQLGQILGQ--PATSADQLVLGPLARG
MSMEG_1143|M.smegmatis_MC2_155 --------SFGGNVGPVGSEQERVTLSVITGQ--PASTATQLLLGPVARG
TH_2386|M.thermoresistible__bu ----------GG--------------------------------------
Mflv_0010|M.gilvum_PYR-GCK AEAPFAPASFGGNVGPVGSQQEQRQLSLITGR--HATPATQLLLGPLARG
Mvan_0959|M.vanbaalenii_PYR-1 --------GYGGNVGPVGSRHELDQLGYITGQ--PATVATQLLLGPVARG
MAV_0102|M.avium_104 --------AYGGNVGPVGSQRERDQLGLITGQGRPASVATQLLLGPVARG
Mb3526c|M.bovis_AF2122/97 ----------GAGPGEHGGGG-----------------------------
Rv3496c|M.tuberculosis_H37Rv ----------GAGPGEHGGGG-----------------------------
*.
MMAR_0177|M.marinum_M AIASRTQQTPTESK--
MUL_4919|M.ulcerans_Agy99 AIASRTQQTPTESK--
MAB_1695|M.abscessus_ATCC_1997 TVVTRAADPHEEGGAR
MSMEG_1143|M.smegmatis_MC2_155 TTVSLAEPTAGEEPR-
TH_2386|M.thermoresistible__bu ----------GEEPR-
Mflv_0010|M.gilvum_PYR-GCK TTVS-----IDGEP--
Mvan_0959|M.vanbaalenii_PYR-1 TTVS-----EGVR---
MAV_0102|M.avium_104 SAVSLQPRPAPGGPK-
Mb3526c|M.bovis_AF2122/97 ----------------
Rv3496c|M.tuberculosis_H37Rv ----------------