For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSAPPAKPKPQRVADPRLRRVLKDDAHKLTAVGGLAALSLDALSSVAYGPEAIVLALIAGGVGAIAFTLP VAIAITVLLIVLVFSYRQVIAVHPEGGGSYAVAKKDLGRGASLVAAASLVVDYVLTVAVSLAAGAASLAS AFPVLAPHLLSITLIGLAILTVINLIGISESAKVLMLPTLLFIACILAVIVFGLMHSTPVAVIGKELGPV EPVSALGIVLVLKAFAAGCSSLTGVEAIANGVPAFKNPAITRAQNTEVALGILLGVMLIGLSVLIKAHHV VPRGDVTILAQLSAAAFGTGLPFYVTNVTVALILGFAANTSFGGLPVLMSLLAKDDRMPHLFGLRAEKPV YRYGVVALSLFSAIILIASDADTHRLLPLFAIGVFIGFTISQTGLVKHWFGARTAGWHWKAALNGFGATL SAIAMVVFFVSKFTEGAWVLLIVIPLLVLLFGRTENYYRSVRSELALDELPTLTPNPPDQTPLVIVPIDD INKRTVQLLEAALHLGGEVVPVTISRTDQRAAAITKRWAQWNPGLELIVLPTKHRSLVTPLVEYVKKRQC DGRQVTVLITEIKTKRRWHQILHNQTGIVLTANLIDRTDAIVANYPYRMN*
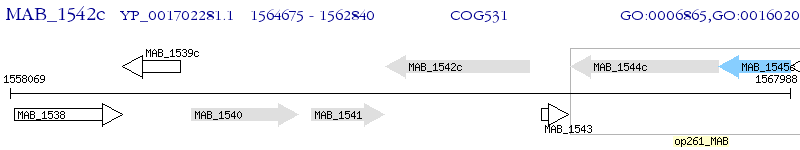
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1542c | - | - | 100% (611) | amino acid permease |
| M. abscessus ATCC 19977 | MAB_2993c | - | 6e-82 | 31.00% (613) | hypothetical protein MAB_2993c |
| M. abscessus ATCC 19977 | MAB_2598 | - | 3e-08 | 23.21% (349) | hypothetical protein MAB_2598 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2709c | - | 8e-80 | 31.38% (615) | hypothetical protein Mb2709c |
| M. gilvum PYR-GCK | Mflv_3926 | - | 1e-79 | 30.83% (613) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv2690c | - | 9e-80 | 31.38% (615) | hypothetical protein Rv2690c |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 3e-06 | 20.61% (427) | putative L-asparagine permease |
| M. marinum M | MMAR_2024 | - | 2e-80 | 31.98% (616) | integral membrane alanine, valine and leucine rich protein |
| M. avium 104 | MAV_3581 | - | 3e-82 | 32.30% (613) | amino acid permease |
| M. smegmatis MC2 155 | MSMEG_2772 | - | 5e-82 | 32.15% (622) | amino acid permease |
| M. thermoresistible (build 8) | TH_2211 | - | 7e-84 | 31.76% (614) | PROBABLE CONSERVED INTEGRAL MEMBRANE ALANINE AND VALINE AND |
| M. ulcerans Agy99 | MUL_3328 | - | 2e-80 | 31.98% (616) | integral membrane alanine, valine and leucine rich protein |
| M. vanbaalenii PYR-1 | Mvan_2474 | - | 5e-79 | 30.51% (613) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3926|M.gilvum_PYR-GCK ---MSKLSTAARRLVLGRPFRSDKLSHTLLPKRIALPVFASDALSSTAYA
Mvan_2474|M.vanbaalenii_PYR-1 ---MSKLSTVARRLVLGRPFRSDRLSHTLLPKRIALPVFASDALSSVAYA
MSMEG_2772|M.smegmatis_MC2_155 ---MSKLSTAARRLVIGRPFRSDKLSHTLLPKRIALPVFASDALSSVAYA
TH_2211|M.thermoresistible__bu ---VSKVSTAARRLLLGRPFRSDRLSHTLLPKRIALPVFASDALSSVAYA
Mb2709c|M.bovis_AF2122/97 ---MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSSIAYA
Rv2690c|M.tuberculosis_H37Rv ---MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSSIAYA
MMAR_2024|M.marinum_M ---MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSSVAYA
MUL_3328|M.ulcerans_Agy99 ---MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSSVAYA
MAV_3581|M.avium_104 ---MSKLSTATRRLLIGRPFRSDRLSHTLLPKRIALPVFASDALSSVAYA
MAB_1542c|M.abscessus_ATCC_199 MVSAPPAKPKPQRVADPRLRRVLKDDAHKLTAVGGLAALSLDALSSVAYG
MLBr_01305|M.leprae_Br4923 ------MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGAI
. .. .* : :. .
Mflv_3926|M.gilvum_PYR-GCK PEEIFLVLSVAGLASYSMAPWIGLAVAAVMLIVIASYRQNVHAYPSGGGD
Mvan_2474|M.vanbaalenii_PYR-1 PEEIFLVLSVAGLAAYSMAPWIGLAVAGVMIIVIASYRQNVHAYPSGGGD
MSMEG_2772|M.smegmatis_MC2_155 PEEIFLVLSVAGLSAYTMTPWIGLAVAAVMLIVIASYRQNVHAYPSGGGD
TH_2211|M.thermoresistible__bu PEEIFVVLSVAGVAAYAMTPWIGLAIATVMALVIASYRHTVQAYPSGGGD
Mb2709c|M.bovis_AF2122/97 PEEIFLVLSVAGLAAYSMAPLIGLAVAAVLLVVVSSYRQNVHAYPSGGGD
Rv2690c|M.tuberculosis_H37Rv PEEIFLVLSVAGLAAYSMAPLIGLAVAAVLLVVVSSYRQNVHAYPSGGGD
MMAR_2024|M.marinum_M PEEIFLMLSVAGLAAYSMAPWIALAVAAVLLVVVSSYRQNVHAYPSGGGD
MUL_3328|M.ulcerans_Agy99 PEEIFLMLSVAGLAAYSMAPWIALAVAAVLLVVVSSYRQNVHAYPSGGGD
MAV_3581|M.avium_104 PEEVFLMLSVAGVSAYALTPWIGLAVAAVMLVVVASYRQNVHAYPSGGGD
MAB_1542c|M.abscessus_ATCC_199 PEAIVLALIAGGVGAIAFTLPVAIAITVLLIVLVFSYRQVIAVHPEGGGS
MLBr_01305|M.leprae_Br4923 GTGLFLGAGGRLAKAGPGLFLVYAVCGVFVFLILRALGELVLHRPSSGSF
:.: : . : . .: ::: : . : *..*.
Mflv_3926|M.gilvum_PYR-GCK YEIATTNLGPTAGLTVASALLVDYVLTVAVSMSSAMANIGSAVPFINQHK
Mvan_2474|M.vanbaalenii_PYR-1 YEVVTTNLGPTAGLTVASALLIDYVLTVAVSMSSAMTNIGSAVPFINDHK
MSMEG_2772|M.smegmatis_MC2_155 YEVVTTNLGPTAGLTVASALLVDYVLTVAVSMSSAMSNIGSAIPFVGHHK
TH_2211|M.thermoresistible__bu YEVVSTNLGRTAGLTVAAALMVDYVLTVAVSISAAMSNIGSVIGFIGEHK
Mb2709c|M.bovis_AF2122/97 YEVVTTNLGATGGLVVASALMVDYVLTVAVSISSAASNIGSVSPFVYEHK
Rv2690c|M.tuberculosis_H37Rv YEVVTTNLGATGGLVVASALMVDYVLTVAVSISSAASNIGSVSPFVYEHK
MMAR_2024|M.marinum_M YEVVSTNLGPTAGLVVASALMVDYVLTVAVSIASAMSNIGSAIPFVYDHK
MUL_3328|M.ulcerans_Agy99 YEVVSTNLGPTAGLVVASALMVDYVLTVAVSIASAMSNIGSAIPFVYDHK
MAV_3581|M.avium_104 YEVVTTNLGETAGLVVASALMVDYVLTVAVSTASAMANIGSAVPIVAEHK
MAB_1542c|M.abscessus_ATCC_199 YAVAKKDLGRGASLVAAASLVVDYVLTVAVSLAAGAASLASAFPVLAPHL
MLBr_01305|M.leprae_Br4923 VSYAREFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFTALPQ
. :* .. ... .:*:.:*. *. :: : : .
Mflv_3926|M.gilvum_PYR-GCK VWFAVAAILVLAALNLRGIRESGTAFAIPTYAFIVGMSVMLIWGFVQIYV
Mvan_2474|M.vanbaalenii_PYR-1 VWFAVIAILVLASLNLRGIRESGTAFAIPTYAFMIGMYAMLAWGFLQILV
MSMEG_2772|M.smegmatis_MC2_155 VLFAVAAILLLAAMNLRGIRESGVAFAIPTYLFMAGMFIMLGWGLFQIYV
TH_2211|M.thermoresistible__bu VWFAVAAILLLAAMNLRGVRESGTAFAVPTYVFVFAMVVMLGWGLIRIYL
Mb2709c|M.bovis_AF2122/97 VLFAVGAIVLIMAMNLRGVRESGLAFAIPTYAFIAGIGTMLVWGLFRIFV
Rv2690c|M.tuberculosis_H37Rv VLFAVGAIVLIMAMNLRGVRESGLAFAIPTYAFIAGIGTMLVWGLFRIFV
MMAR_2024|M.marinum_M VFFAVAAILLVTALNLRGVRESGLAFAIPTYAFIIGISAMLGWGLFRIYV
MUL_3328|M.ulcerans_Agy99 VFFAVAAILLVTALNLRGVRESGLAFAIPTYAFIIGISAMLGWGLFRIYV
MAV_3581|M.avium_104 VFFCVAAILLVMALNLRGVRESGVAFAIPTYAFIIGVLTMIGWGLFRIFV
MAB_1542c|M.abscessus_ATCC_199 LSITLIGLAILTVINLIGISESAKVLMLPTLLFIACILAVIVFGLMHSTP
MLBr_01305|M.leprae_Br4923 WTLALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAFLVVGTIFLGG
: : .: :: :** .: . .: : : .: * .
Mflv_3926|M.gilvum_PYR-GCK LGTPLRAESADFEMHSEHGDVLGFALIFLVARA----FSSGSAALTGVEA
Mvan_2474|M.vanbaalenii_PYR-1 LGRDLRAESAGFEMHSEHGEVLGFALVFLVARA----FSSGSAALTGVEA
MSMEG_2772|M.smegmatis_MC2_155 LDHSLRAESADFQMHSEHGEVLGFALVFLVARA----FSSGSAALTGVEA
TH_2211|M.thermoresistible__bu FDTPLRAESAAFEMIPPDGEIVGFALVFLVARA----FSSGSAALTGIEA
Mb2709c|M.bovis_AF2122/97 LGNPVRAESAAFEMHAEHGQIVGFALVFLVARS----FSSGCAALTGVEA
Rv2690c|M.tuberculosis_H37Rv LGNPVRAESAAFEMHAEHGQIVGFALVFLVARS----FSSGCAALTGVEA
MMAR_2024|M.marinum_M LGNQVRAESAGFVMHAEHGKVVGIALVFLVARS----FSSGCAALTGVEA
MUL_3328|M.ulcerans_Agy99 LGNQVRAESAGFVMHAEHGKVVGIALVFLVARS----FSSGCAALTGVEA
MAV_3581|M.avium_104 LGNPLRAESAGFEMHAQHGQVVGFALAFLVARS----FSSGCAALTGVEA
MAB_1542c|M.abscessus_ATCC_199 VAVIG-KELGPVEPVSALG-------IVLVLKA----FAAGCSSLTGVEA
MLBr_01305|M.leprae_Br4923 R-YPVDGHNTGLSLWTSHGGLFPTGVAPLIVVSSGVMFAYAAVELVGTAA
. . . * *: : *: .. *.* *
Mflv_3926|M.gilvum_PYR-GCK ISNGVPAFRKPKSRNAATTLLLLGLIATTLLMGIIMLARAIGVQIAERPA
Mvan_2474|M.vanbaalenii_PYR-1 ISNGVPAFRKPKSRNAATTLLLLGVIAITLFMGMILLARATGVQVAERPL
MSMEG_2772|M.smegmatis_MC2_155 ISNGVPAFRKPKSRNAATTLLLLGMIAVTLFMGIILLAKATGVQIAEHPH
TH_2211|M.thermoresistible__bu ISNGVPAFRKPKARNAATTLLVLGLIAVPMFLGTVVLAERTGVQIAERPH
Mb2709c|M.bovis_AF2122/97 ISNGVPAFQKPKSRNAATTLLMLGIIAVSMFMGMIVLAVETGVQVVDDPD
Rv2690c|M.tuberculosis_H37Rv ISNGVPAFQKPKSRNAATTLLMLGIIAVSMFMGMIVLAVETGVQVVDDPD
MMAR_2024|M.marinum_M ISNGVPAFQKPKSRNAATTLLMLGAIAVALLMGVVALAVQTGAQVVDDPD
MUL_3328|M.ulcerans_Agy99 ISNGVPAFQKPKSRNAATTLLMLGAIAVALLMGVVALAVQTGAQVVDDPD
MAV_3581|M.avium_104 ISNGVPAFQKPKSRNAATTLLMLGGIAVTLLMGIIVLAEKIGVKLVEDPA
MAB_1542c|M.abscessus_ATCC_199 IANGVPAFKNPAITRAQNTEVALGILLGVMLIGLSVLIKAHHVVPRGD--
MLBr_01305|M.leprae_Br4923 GETVEPKKIMPRAINSVIARIAIFYVGSVILLALLLPYSAFKAS------
. * * .: : : : : :::. .
Mflv_3926|M.gilvum_PYR-GCK EQLVGAPEDYHQKTLIAQLAETVFHGFPFGLFFIALVTALILVLAANTAF
Mvan_2474|M.vanbaalenii_PYR-1 EQLGGAPADYQQKTLIAQLADAVFHDFPLGLYLIAGVTALILVLAANTAF
MSMEG_2772|M.smegmatis_MC2_155 VQLIGAPTDYHQKTLIAQLADAVFHGFPLGLYLIATVTALILCLAANTAF
TH_2211|M.thermoresistible__bu AQFVGAPPDYQQKTLIAQLAGAVFDGWGPAVLATVIAAALILLLAANTAF
Mb2709c|M.bovis_AF2122/97 TQLTGAPPGYQQKTLVAQLAQAVFGGFYLGFLLIAAVTALILVLAANTAF
Rv2690c|M.tuberculosis_H37Rv TQLTGAPPGYQQKTLVAQLAQAVFGGFYLGFLLIAAVTALILVLAANTAF
MMAR_2024|M.marinum_M TQLTGAPDGYQQKTLVAQLAQAVFGSFHLGFLMIAAVTALILVLAANTAF
MUL_3328|M.ulcerans_Agy99 TQLTGAPDGYQQKTLVAQLAQAVFGSFHLGFLMIAAVTALILVLAANTAF
MAV_3581|M.avium_104 TQLSGTPPGYYQKTLVTQLAQAVFGGFHIGFLLIATVTALILVLAANTAF
MAB_1542c|M.abscessus_ATCC_199 ------------VTILAQLSAAAFG-TGLPFYVTNVTVALILGFAANTSF
MLBr_01305|M.leprae_Br4923 -----------ESPFVTFFSKVGFYGAGDLMNIVVLTAALSSLNAGLYAT
.::: :: . * . ..** *. :
Mflv_3926|M.gilvum_PYR-GCK NGFPVLGSILAQDRFLPRQLHTRGDRLAFSNGILFLAFGAVAFVVAFQAE
Mvan_2474|M.vanbaalenii_PYR-1 NGFPVLGSILAQDRFLPRQLHTRGDRLAFSNGILFLAFGAIAFVVAFQAE
MSMEG_2772|M.smegmatis_MC2_155 NGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGILILALAAIAFVVAFGAE
TH_2211|M.thermoresistible__bu NGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGILFLSLVASAFVVAFGAE
Mb2709c|M.bovis_AF2122/97 NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGILFLAAAAIGAVVAFRAE
Rv2690c|M.tuberculosis_H37Rv NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGILFLAAAAIGAVVAFRAE
MMAR_2024|M.marinum_M NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGIIFLSAAAVGAVVAFRAE
MUL_3328|M.ulcerans_Agy99 NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGIIFLSAAAVGAVVAFRAE
MAV_3581|M.avium_104 NGFPVLGSILAQHSYLPRQLHTRGDRLAFSNGILFLSGAALLAIIAFRGE
MAB_1542c|M.abscessus_ATCC_199 GGLPVLMSLLAKDDRMPHLFGLRAEKPVYRYGVVALSLFSAIILIASDAD
MLBr_01305|M.leprae_Br4923 G--RVMHSIAINGS--GPKFTARMSKNGVPYGGILLAAVICLCGVALNAF
. *: *: : : * .: * : *: :* .
Mflv_3926|M.gilvum_PYR-GCK VTKLIQLYIVGVFVSFTLSQIGMVRHWTRLLRTETDAAARGRMVRARVIN
Mvan_2474|M.vanbaalenii_PYR-1 VTALIQLYIVGVFVSFTFSQIGMVRHWTRLLRTETDPAARRRMMRARVIN
MSMEG_2772|M.smegmatis_MC2_155 VTALIHLYIVGVFVSFTFSQIGMVRHWTRLLRNETDPAIRRRMMRSRAIN
TH_2211|M.thermoresistible__bu VSALIQLYIVGVFVSFTFSQLGMVRHWTRSLRGETEPAVRRTMHRARAVN
Mb2709c|M.bovis_AF2122/97 LTALIQLYIVGVFISFTMSQVGMVRHWTRLLSAETDPRARRAMLRSRAVN
Rv2690c|M.tuberculosis_H37Rv LTALIQLYIVGVFISFTMSQVGMVRHWTRLLSAETDPRARRAMLRSRAVN
MMAR_2024|M.marinum_M LTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDVAARRKMVRSRVVN
MUL_3328|M.ulcerans_Agy99 LTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDVAARRKMVRSRVVN
MAV_3581|M.avium_104 VTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDPAARRKMMRSRAVN
MAB_1542c|M.abscessus_ATCC_199 THRLLPLFAIGVFIGFTISQTGLVKHWFGARTAG--------WHWKAALN
MLBr_01305|M.leprae_Br4923 NPG------QAFEIVLSVAALGIIAGWG----------------------
.. : ::.: *:: *
Mflv_3926|M.gilvum_PYR-GCK TVGFLCTGTVLIIVVVTKFVVGAWIAILAMGALFGIMKLIHRHYASVSRE
Mvan_2474|M.vanbaalenii_PYR-1 SVGLVCTGTVLVIVVVTKFLAGAWIAILAMSALFGLMKLIHKHYASVTRE
MSMEG_2772|M.smegmatis_MC2_155 TVGLTATGAVLVVVVVTKFVAGAWIAILAMVALFVLMKLIHRHYDTVARE
TH_2211|M.thermoresistible__bu AVGCVATGSVLLVVVVTKFAAGAWMAILTMAVLFMLMRLIHRHYATVARE
Mb2709c|M.bovis_AF2122/97 TVGFVSTGTVLLIVLVTKFLAGAWIAIVAMGGFFMMMKLIHRHYDAVNRE
Rv2690c|M.tuberculosis_H37Rv TVGFVSTGTVLLIVLVTKFLAGAWIAIVAMGGFFMMMKLIHRHYDAVNRE
MMAR_2024|M.marinum_M SVGLVSTGAVLLIVLVTKFLAGAWIAIVAMGSVFILMKMIRRHYDTVNRE
MUL_3328|M.ulcerans_Agy99 SVGLVSTGAVLLIVLVTKFLAGAWIAIVAMGSVFILMKMIRRHYDTVNRE
MAV_3581|M.avium_104 TVGLLSTGAVLLVVLITKFAAGAWIALFAMSLLFGIMKMINRHYNTVNRE
MAB_1542c|M.abscessus_ATCC_199 GFGATLSAIAMVVFFVSKFTEGAWVLLIVIPLLVLLFGRTENYYRSVRSE
MLBr_01305|M.leprae_Br4923 -------TIVLCQLRLHKMAKAGIMRRPRFR-------------------
.: . : *: .. : :
Mflv_3926|M.gilvum_PYR-GCK LEARAADAED-IVLPSRNHAVVLVSNLHMPTLRALAYARATRPDVLEAIT
Mvan_2474|M.vanbaalenii_PYR-1 LEARAAETED-IVLPSRNHAIVLVSNAHLPTLRALAYARATRPDVLEAIT
MSMEG_2772|M.smegmatis_MC2_155 LEAQAEDEDD-IVLPSRNHAVVLVSKLHLPTKRALAYARATRPDVLEAIT
TH_2211|M.thermoresistible__bu LDERAAADDDGMVLPSRNHAIVLVSNLHLPTLRALAYARATRPDVLEAVT
Mb2709c|M.bovis_AF2122/97 LAEQAEEA--EITLPSRNHAVVLVSKLHLPTLRALTYARATRPDVLEAVT
Rv2690c|M.tuberculosis_H37Rv LAEQAEEA--EITLPSRNHAVVLVSKLHLPTLRALTYARATRPDVLEAVT
MMAR_2024|M.marinum_M LEEQAAAHDNEVVLPSRNHALVLVSKLHLPTLRALAYARATRPDVLEAIT
MUL_3328|M.ulcerans_Agy99 LEEQAAAHGNEVVLPSRNHALVLVSKLHLPTLRALAYARATRPDVLEAIT
MAV_3581|M.avium_104 LAEQAADQD-DVVLPSRNHAVVLVSKLHLPTLRALAYARATRPDVLEAVT
MAB_1542c|M.abscessus_ATCC_199 LALDELPTLTPNPPDQTPLVIVPIDDINKRTVQLLEAALHLGGEVVP-VT
MLBr_01305|M.leprae_Br4923 -------------MPLAPYSGYLTLAFLFAVLVVMAFDKPIGTWTVASLI
. : .: :
Mflv_3926|M.gilvum_PYR-GCK VSVDDAETRELVHKWENSDVSVPLKVIASPYREITRPVLDYVKRVTKDSP
Mvan_2474|M.vanbaalenii_PYR-1 VSVDDAETRQLVHKWEHSDISVPLKVVASPYREITRPVLDYVKRVGKDSP
MSMEG_2772|M.smegmatis_MC2_155 VSVDDAETRALVHEWEGSDVAVPLKVIASPYREITRPVLDYVKRITKESP
TH_2211|M.thermoresistible__bu VSVDDAETRELVHKWEESDVTVPLKVIASPYREVTRPVLDYVKRVTKESP
Mb2709c|M.bovis_AF2122/97 VNVDDAETRELVRQWQDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
Rv2690c|M.tuberculosis_H37Rv VNVDDAETRELVRQWQDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
MMAR_2024|M.marinum_M VSVDDVETRELVRQWEDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
MUL_3328|M.ulcerans_Agy99 VSVDDVETRELVRQWEDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
MAV_3581|M.avium_104 VSVDDAETRDLVHKWEEADISVPLKVIASPYREITRPVLEYVKRAREESP
MAB_1542c|M.abscessus_ATCC_199 ISRTDQRAAAITKRWAQWNPGLELIVLPTKHRSLVTPLVEYVK--KRQCD
MLBr_01305|M.leprae_Br4923 VIVPALIAGWYSIRKRVMTIARERMGYTGPFPAIANPPVQPSERSHSQNP
: : . . . :. * :: : :
Mflv_3926|M.gilvum_PYR-GCK RTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFEPNVMVTSVPWQLTS
Mvan_2474|M.vanbaalenii_PYR-1 RTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFEPNVMVTSVPWQLSS
MSMEG_2772|M.smegmatis_MC2_155 RTVVTVFIPEYVVGHWWEQILHNQSALRLKGRLLFMPNVMVTSVPWQLTS
TH_2211|M.thermoresistible__bu RTVVTVFIPEYVVGHWWEQLLHNQSALRLKGRLLFMPNVMVTSVPWQLSS
Mb2709c|M.bovis_AF2122/97 RTVVTVFIPEYVVGRWWEQLLHNQSALRLKGRLLFMPGVMVTSVPWQLTS
Rv2690c|M.tuberculosis_H37Rv RTVVTVFIPEYVVGRWWEQLLHNQSALRLKGRLLFMPGVMVTSVPWQLTS
MMAR_2024|M.marinum_M RTVVTVFIPEYVVGRWWEQLLHNQSAFRLKTRLLFMPGVMVTSVPWQLTS
MUL_3328|M.ulcerans_Agy99 RTVVTVFIPEYVVGRWWEQLLHNQSAFRLKTRLLFMPGVMVTSVPWQLTS
MAV_3581|M.avium_104 RTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFMPGVMVTSVPWQLSS
MAB_1542c|M.abscessus_ATCC_199 GRQVTVLITEIKTKRRWHQILHNQTGIVLTANLIDRTDAIVANYPYRMN-
MLBr_01305|M.leprae_Br4923 --------------------------------------------------
Mflv_3926|M.gilvum_PYR-GCK SERLKKMAPQSAPGDARRGFLE
Mvan_2474|M.vanbaalenii_PYR-1 SERLKKLAPQSAPGDARRGFLD
MSMEG_2772|M.smegmatis_MC2_155 SERLKAIQRQSAPGDARRGFLE
TH_2211|M.thermoresistible__bu SERRRKLQPQWAPGDARRGFLE
Mb2709c|M.bovis_AF2122/97 SERIKTLQPHAAPGDTRRGIFD
Rv2690c|M.tuberculosis_H37Rv SERIKTLQPHAAPGDT------
MMAR_2024|M.marinum_M SERINTLEPHAAPGDMRRGIFD
MUL_3328|M.ulcerans_Agy99 SERINTLEPHAAPGDTRRGIFD
MAV_3581|M.avium_104 SERRKTLEPHMAPGDARRGIFD
MAB_1542c|M.abscessus_ATCC_199 ----------------------
MLBr_01305|M.leprae_Br4923 ----------------------